Abstract
OBJECTIVE To investigate homozygous deletions and mutations in the CDKN2A gene (p16INK4a and p14ARF gene) in hydatidiform moles.
METHODS A total of 38 hydatidiform mole samples and 30 villi samples were examined for homozygous deletions in the CDKN2A gene by PCR and for mutations by DHPLC.
RESULTS i)Among 38 hydatidiform mole samples, homozygous deletions in the p16INK4a exon 1 were identified in 5 cases (13.2%), while no homozygous deletions were found in the p16INK4a exon 1 of 30 early-pregnancy samples. The rates of those deletions in hydatidiform compared to early-pregnancy villi samples was statistically significant (P = 0.036). ii) No homozygous deletions in the p14ARF exon 1 or p16INK4a exon 2 were found in any of the hydatidiform moles or early-preganancy samples. iii) In all hydatidiform moles and early-pregnancy villi samples, no mutations were detected by DHPLC.
CONCLUSION We suggest there may be a close correlation between homozygous deletions in the CDKN2A gene and occurrence of hydatidiform moles variation in the CDKN2A gene is mainly caused by homozygous deletions, while mutations may be not a major cause.
keywords
Introduction
Hydatidiform moles are the most common gestational trophoblastic disease. They are formed by excessive proliferation of gestational trophoblastic cells of the placental villi, and are prone to invasion elsewhere. The cyclin-dependent kinase inhibitor gene (CDKN2A), which includes the p14ARF and p16INK4a genes, is a tumor suppressor gene with genetic variations that have been found in many tumors in previous studies. This study was designed to detect homozygous deletions in the of CDKN2A gene in hydatidiform moles using the polymerase chain reaction (PCR) and to detect mutations by denaturing high performance liquid chromatography (DHPLC). We also aimed to find potential relationships between variations in the CDKN2A gene and occurrence of hydatidiform moles.
Materials and Methods
Subjects
The study was approved by the ethics committee of Peking University Third Hospital. Informed consent was obtained from all patients. A total of 38 hydatidiform mole villi samples and 30 early-pregnancy villi samples were included in our study. The tissues were collected from Peking University Third Hospital, Beijing Haidian Maternal and Child Health Hospital and Affiliated First Hospital of Liaoning Medical College.
DNA extraction
Early-pregnancy villi samples and typical hydropic vesicles of hydatidiform mole villi tissues were washed 3 times in a precooled NaCl solution in order to remove any possible blood. Then the samples were ground in a tissue grinder with 3~4 volumes of liquid which included 20 mmol/L Tris, 100 mmol/L NaCl and 50 mmol/L EDTA (pH 7.5) to produce a cell suspension to which proteolytic enzyme K and SDS were subsequently added. Genomic DNA was extracted using phenol and chloroform after which it was dissolved in TE (10 mmol/L Tris-HCl, 1 mmol/L EDTA) liquid followed by freezing at -20°C until use.
PCR primers
Primers for the p14ARF exon 1 and β-actin gene were synthesized according to previous literature[1] by the Beijing Augct Technologic Co. which also designed and synthesized primers for the p16INK4a exon 1 and 2. The primer sequences were as follows: β-actin (193 bp) sense: 5’-GCA TCC ACG AAA CTA CCT TC-3’;; antisense: 5’-TCA GCT CAG GCA GGA AAG AC-3’ p14ARF exon 1 (149 bp) sense: 5’-GAG TGA GGG TTT TCG TGG TT-3’; antisense: 5’-GCC TTT CCT ACC TGG TCT TC-3’. p16INK4a exon 1 (384 bp) sense: 5’-TAG CAC CTC CTC CGA GCA CTC-3’; antisense: 5’-CAG ACC CTC TAC CCA CCT GGA-3’. p16INK4a exon 2 (386 bp) sense: 5’-CCT GGC TCT GAC CAT TCT GTT C-3’; antisense: 5’-GCT TTG GAA GCT CTC AGG GTA C-3’.
PCR amplication of the p14ARF exon 1, β-actin, and p16INK4a exon 1 and 2
The total reaction volume was 25 μl and amplication parameters were set as follows: pre-degeneration for 5 min at 94°C, degeneration for 30 s at 94°C, annealing for 30 s at 57°C, elongation for 30 s at 72°C with 35 cycles and final elongation for 7 min at 72°C. Then the PCR amplified products were examined by 1.5% agarose gel electrophoresis.
DHPLC detection
DHPLC detection was performed using the WAVEDNA fragment analysis system (Transgenomic, Santa Clara, CA). Before conducting the DHPLC procedure, amplified products were mixed with a reference PCR product (wild-type). Then the mixture was subjected to a 3-min 95°C denaturing step followed by gradual reannealing from 95°C to 45°C within a period of 30 min to form potential heteroduplexes. The mobile phase was a 0.1 M triethylammonium acetate buffer at pH 7.0 (TEAA), also containing 0.1 mM EDTA. Then the mixture was eluted with a linear acetonitrile gradient of 1.8% per min at a flow-rate of 0.9 ml/min. The start- and end-points of the gradient were adjusted according to the size of the PCR products. Generally, analysis took less than 8 min, including column regeneration and re-equilibration to the starting conditions. The temperature required for successful resolution of the heteroduplex molecules was determined empirically by injecting PCR products at increasing mobile phase temperatures until a significant decrease in retention was observed. At this point, at least 95% of all mismatches can be detected, with further improvements in resolution.
Results
PCR amplication of the p14ARF exon 1, β-actin, and p16INK4a exon 1 and 2
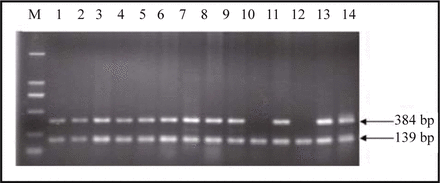
Serving as a control, β-actin was amplified along with all 38 hydatidiform mole and 30 villi samples. No homozygous deletions in the p14ARF exon 1, p16INK4a exon 1 or 2 were detected in all 30 early-pregnant villi samples. There were no homozygous deletions in the p14ARF exon 1 or p16INK4a exon 2 in 38 hydatidiform mole samples, while the p16INK4a exon 1 products of the 5 hydatidiform mole samples failed to be detected by agarose gel electrophoresis (Fig.1). The homozygous deletion rate in the p16INK4a exon 1 between the hydatidi-form mole and early-pregnant villi samples was statistically significant (P = 0.036).
P16INK4a exon 1 amplified products of hydatidiform mole samples numbered as 10 and 12 failed to be detected while their β-actin products were well amplified.
DHPLC analysis
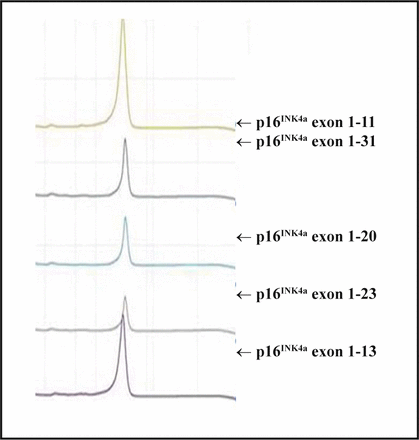
After DHPLC analysis, all spectra of the PCR amplication products of the p14ARF exon 1 and p16INK4a exon 1 and 2 were single peak curves which indicated that the sequences of the amplified products were coincident with the wild-type, and no mutations existed (Fig.2).
DHPLC detection for p16INK4a exon 1 amplication products.
Discussion
The human CDKN2A gene which is located on chromosome 9p21 is a cyclin-dependent kinase inhibitor gene that encodes 2 different tumor suppressor proteins. One is designated as p16INK4a which is encoded within exon 1α, exon 2 and 3. The other is designated as p14ARF and is encoded within exon 1β, exon 2 and 3. The p16INK4a and p14ARF genes share exon 2 and 3, while they have different open-reading frames and their amino sequences are quite disparate. The CDKN2A gene regulates cell proliferation by 2 different pathways[2]: p16INK4a-CDK4/CDK6-pRb and p14ARF-mdm2-p53. Variation of the gene or inactivation of the protein will result in loss of control of regulatory pathways, excessive cell proliferation and induction of tumorigenesis[3-5].
DHPLC is a sensitive technique for mutation detection[6,7]. The technique separates heteroduplex from homoduplex molecules by ion-pair reverse-phase liquid chromatography and, since it is automated, renders screening for mutations in a less expensive and timely manner. The power of the approach is especially realized in those instances when sequence variation is low, and consequently only homoduplex patterns are typically observed[8,9]. In our study, all spectra of PCR products of the p14ARF exon 1 and p16INK4a exon 1 and 2 were single peak curves, indicating that sequences of the amplified products were coincident with the reference (wild-type) sequences and no mutations existed.
Homozygous deletions in the p16INK4a exon 1 were detected in 5 hydatidiform mole cases in our study. These genetic variations may result in disordered regulation, abnormal cell division and excessive proliferation, suggesting that the genetic variation of the CDKN2A gene may be related to proliferation of the placental hydatidi-form moles. Wang et al.[10] reported that deletion rates of the p16 gene and p15 gene and their proteins were significantly higher in hydatidiform moles compared to in villi samples. In 2004, Xue et al.[11] detected hypermethylation of genes (p16, HIC-1, TIMP-3, GSTP1, DAPK, E-cadberin) in hydatidiform mole and chorioepithelioma samples using methylation specific PCR (MSP), and found that hypermethylation of the p16 gene showed a highly correlated relationship with the progression of hydatidiform moles.
Variations in the p16INK4a/p14ARF genes, including homozygous deletion, heterozygous deletion, mutation and promoter hypermethylation, have been found in many kinds of tumor malignancies. Pineau et al.[12] detected homozygous deletions in 246 gene sites in lung cancer, ovarian cancer and squamous cell carcinoma of the head and neck, and found that homozygous deletions of p16INK4a/p14ARF were the most common event.
In 2005, Grafstrom et al.[13] studied the relationship between homozygous deletions of the INK4 gene in 86 malignant melanoma patients and their prognosis, and found that these deletions were the most important genetic change which correlated to poor prognosis. Using Southern blots, PCR-single-strand conformation polymorphism (PCR-SSCP), MSP and reverse transcription PCR (RT-PCR), Chang et al.[14] detected variations in the p16INK4a/p14ARF gene, and discovered that most deletions occurred in exon 2, and that promoter hypermethylation took place in 60% of the cases. All the above studies indicated that variations in the p16INK4a/p14ARF gene existed in numerable tumors and related to their progression.
In conclusion, our results indicated that homozygous deletions in the CDKN2A gene were highly related to the occurrence of hydatidiform moles, while mutation may be not the major cause for its genetic variation. Combined with previous studies, we suggest that homozygous deletions and promoter hypermethylation of the CDKN2A gene may be the major cause for its variations in hydatidiform moles. Further studies with larger number of cases should be conducted to explain the occurrence of these tumors, and to elucidate their development.
Footnotes
This work was supported by a grant from the National Natural Science Foundation of China (No. 30772321).
- Received November 16, 2007.
- Accepted January 10, 2008.
- Copyright © 2008 by Chinese Anti–Cancer Association