Abstract
OBJECTIVE It has been shown that application of molecular biological techniques to surgical margins of some cancers could predict risk of local recurrence. However, the optimal length of surgical resection with tumor-free surgical margins for esophageal squamous cell carcinoma (ES-CC) Is unknown. This study was conducted to evaluate the optimal length of surgical resection for ESCC with molecularly tumor-free surgical margins marked by p53 and Ki67.
METHODS Surgical specimens from 70 patients with ESCC were collected for study. The lengths of the upper margin, tumor, and lower margin of every specimen were measured during the operation. Each specimen was divided Into three large pathologic sections, stained with H&E and Immunohistochemlcally for p53 and KI67, and examined microscopically. The lengths of the upper and lower resection ends were measured for p53 and KI67 positive expression. The actual surgical lengths were calculated by the principle of rational shrinkage.
RESULTS All surgical margins were histologically tumor-free, while the positive rates of p53 and KÍ67 were 66% and 54%. The positive rates of p53 and KI67 In the upper resection end were 17% and 20%. The mean lengths of the upper resection end showing p53 and KI67 positive expression were 1.08 ±1.12 cm and 1.64 ±1.01 cm, and the maximum lengths were 3.73 cm and 3.26 cm. The positive rates of p53 and KI67 in the lower resection end were 20% and 23%. The mean lengths of the lower resection end of p53 and KI67 with positive expression were 1.11 ±1.15 cm and 1.34±0.94 cm, and the maximum lengths were 3.73 cm and 3.61 cm.
CONCLUSION The optimal length of surgical resection with molecularly tumor-free surgical margins of ESCC is not more than 5 cm.
keywords
Esophageal cancer is the sixth most common cause of cancer-related death worldwide.[1] China is one of the countries with a high incidence of esophageal cancer, and more than 85 percent of esophageal cancer in China is squamous cell carcinoma.[2,3] The incidence of esophageal adenocarcinoma has increased from the 1970's in the United States and Europe.[4,5]
Traditionally, histopathological hematoxylin and eosin (H&E) staining is used to histologically assess resection margins in resections of esophageal carcinoma. The “molecular resection margin” concept was introduced into surgical oncology with the development of molecular biology. Genetic and epigenetic alterations are hallmarks of human cancers, including proto-oncogene activation and suppressor gene inactivation via mutations, or loss, and promoter hypermethylation.[6] Since genetic alterations precede phenotypic changes, histologic assessment alone may be inadequate to accurately detect the presence of transformed cells in resection margins.[7]
Brennan et al. [8] put forward the “molecular resection margin” concept for the first time when they used the polymerase chain reaction (PCR) to detect p53 gene mutations in the resection margin tissues of the head and neck. They found that even if the resection margins were negative using traditional histopathological H&E staining, the rate of local recurrence was high in those patients whose resection margins were positive using molecular analysis.
Shao et al. [9] found similar results in studies of esophageal squamous cell carcinoma. They used PCR to detect p53 gene mutations in 23 patients with esophageal squamous cell carcinoma whose resection margins were negative by histopathological examination. In 13 cases of these with p53 gene mutations in the resection margin tissue, these were followed for 3—16 months after operation, 6 cases recurred. No cases recurred among 10 cases without p53 gene mutations. Masasyesva et al.[10] also found that molecular margin analysis predicted local recurrence after sublobor resection of lung cancers. Other researchers have reported similar results suggesting that the rate of local recurrence was higher in those patients whose molecular resection margin was positive.[11,12]
The above studies demonstrated that the patients with positive molecular margins have more local recurrences even when their histopathological resection margins were negative. A significant limitation of these molecular resection margin studies is they just focused on the relationship between local recurrence and a positive molecular margin. Surgeons also need to know how they can achieve a margin that is negative by both histopathology and molecular analysis. Does the traditional 5-centimetre-margin for esophageal squamous cell carcinoma still achieve a clean molecular resection margin? What is the optimal resection length to make sure that the molecular resection margin is clean for esophageal squamous cell carcinoma? This study was designed to answer these questions.
The p53 gene,is a well-known tumor suppressor gene,that is believed to serve as a gatekeeper against carcinogenesis.[13] Under normal circumstances.the function of the p53 protein is to prevent the propagation of genetically damaged cells.p53 assists in DNA repair by causing G1 arrest and inducing DNA repair genes or directs apoptosis in cells,which are genetically damaged beyond repair.[13] Cells with a loss of p53 function are believed to undergo malignant transformation.[13] Gao et a1.[14] demonstrated that p53 protein accumulation and gene mutation may occur at very early stages of esophageal carcinogenesis. In previous studies, the p53 gene has been used the most often as a molecular-resection margin marker. We continued to use the p53 gene as a molecular margin marker in our study.
Ki67 is a nuclear non-histone protein expressed maximally in cells in the G2 and M phases of the cell cycle, but absent in resting cells.[15] Hence, Ki67 can be employed to measure the growth fraction of normal tissues and malignant tumors.[16] Xu et al.[17] showed that expression of p53 and Ki67 in the normal, premalignant and malignant esophageal tissues has obvious differences. We chose Ki67 as another molecular resection marker in our research.
In summary, the aim of this study was to determine the optimal molecular resection margin length for esophageal squamous cell carcinoma. We choose p53 and Ki67 as molecular resection margin markers and large pathologic sections as a methodology in this study.
MATERIALS AND METHODS
Patient information
Specimens from 70 patients with esophageal squamous cell carcinoma, who underwent resections at the Cancer Center of the Sun Yat-sen University between July, 1999 to December, 2000, were collected for study. All the histopathological resection margins in both upper and lower ends were negative. There were tissues from 53 males and 17 females. The sites of tumors were upper thoracic in 12; middle thoracic in 47 and lower thoracic in 11. Three patients were in Stage I, 35 in Ha, 4 in. Hb and 28 in HI. The histopathological Grades were as follows: Grade I, 37; II, 26; and III, 7.
Length of specimen measurement
During the operation, the length of the upper margin, tumor, and lower margin were measured before the esophagus was cut. After the esophagus with the tumor was removed, specimens were opened longitudinally on the side opposite the tumor. The length of the upper margin, tumor and lower margin were measured again. A large pathologic section was taken and the length of upper margin, tumor and lower margin of the large pathologic sections was measured for the third time.
Methods to treat large pathologic sections
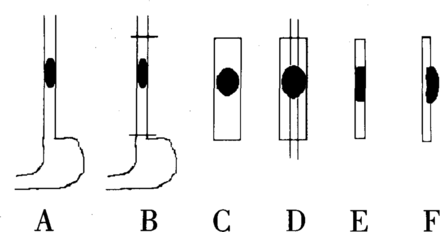
The technique of treating the large pathologic specimens as been previously reported.[17] In brief: each specimen was pinned onto a corkboard. After formalin fixation for one week, the whole esophagus with the tumor mass was sliced into a longitudinal strip of about 5 mm thick. They were composed of the maximal longitudinal part of the tumor, a part of normal esophagus proximal and distal to the tumor. The specimen was then dehydrated using 60%, 70%, 80% and 95% alcohol in turn each for 24 h and twice in 100% alcohol for 2 h. Hyalinization was conducted by placeing the specimen into clove oil for 1 h and into dimethylbenzene for 6 h. The specimens were then placed in paraffin blocks and sliced. The large pathologic section was sliced to 10 |xm sections, and the slices put into water, and then placed onto a large glass slide. (Figs. 1, 2).
Process of making an esophageal large pathologic section.
A: Site of the tumor, B: Resection of the esophagus, C: Specimens were opened longitudinally, D: A longitudinal strip about 5 mm thick was sliced, E: A large pathologic paraffin block was made, F: Slice of large pathologic section.
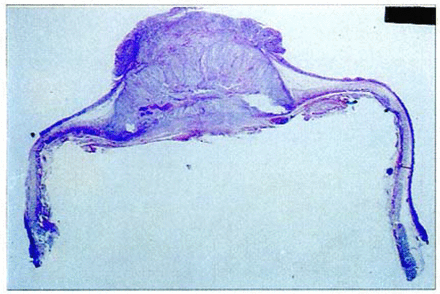
Photo of a large pathologic section of esophageal squamous cell carcinoma. The large pathologic section included the esophageal tumor, the upper mucosa and lower mucosa of the esophagus.
H&E and immunohistochemical (IHC) staining
Each specimen was divided into three large pathologic sections. One large pathologic section was stained with H&E, the other two sections were stained for p53 and Ki67. The monoclonal antibody used in this series was mouse monoclonal primary anti-p53 antibody (DAKO, dilution 1:100) and rabbit monoclonal anti-Ki67 antibody (DAKO, dilution 1:80) from the DAKO Corp. (Carpintería, CA). The slides were viewed and photographed under a light microscope. Controls were established by replacing the primary antibody with PBS and normal mouse serum. Known immunostaining-positive slides were used as positive controls.
Scoring of Ihe p53 and Ki67 staining
The evaluation of the immunohistochemistry was done by two pathologists blinded to the clinical and pathologic characteristics of the patients. Staining for p53 or Ki67 was scored as +++ if more than 50% of the cells were immunostained positive; ++ if 26% to 50% of cells were positive; + if 11% to 25% of cells were positive; and - if less than 10% of cells were positive. The p53 status of the tumor and mucous layer of the upper and lower ends, Ki67 status of tumor and mucous layer of the upper and lower ends, were observed and measured.
Calculations of the actual length during operations
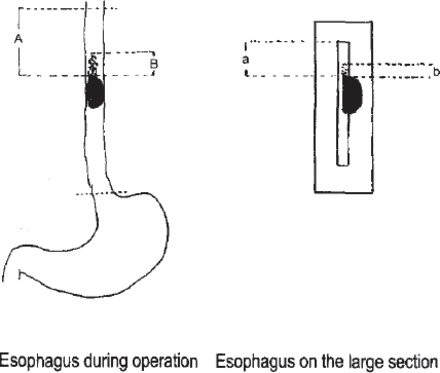
Due to the shinkage of the esophagus during the process of making the large slice, the length of the items under the microscope was not the actual length previously measured during the operation.[18, 19] We calculated the actual length during the operation by the principle of rational shrinkage (Fig.3).
Extrapolationactual length during the operation. A: Proximal surgical resection length during the operation, B: Notional safe margin length, a: Proximal surgical resection length of the large pathologic section, b: Length of positive expression of the large pathologic section. According to the principle of relative shrinkage: B/A=b/a. We can calculate the length of at operation: B=Axb/a.
Follow up methods
Follow-up started after surgery and continued until the end of the study or death of the patient. Data from all patients who died from other causes or were lost to follow-up were censored.
RESULTS
Tumor positive expression
Of the total 70 patients with ESCC, 46 (66%) showed positive expression of p53 (Table 1) and 38 (54%) expressed Ki67 (Table 2).
p53 Expression
Ki67 Expression
Length of upper end positive expression
In the mucous layer of the upper end, 12 patients showed positive expression of p53 and 14 patients showed positive expression of Ki67 (Tables 1, 2). Most p53 positive expression lengths were less than 2.0 cm (Table 3). Most Ki67 positive expression lengths were less than 3.0 cm (Table 3). The longest lengths of positive expression of p53 and i67 were 3.73 cm and The longest lengths of positive expression of p53 and Ki67 were 3.33 cm and 3.26 cm (Table 4).
Length distribution of p53 and Ki67-positive expression beyond tumor
Length of p53 and Ki67-positive expression beyond tumor (cm)
Lengths of lower end positive expression
In the mucous layer of the lower end, 14 patients displayed positive expression of p53 and 16 patients had positive expression of Ki67 (Tables 1, 2). Most p53 positive expression lengths were less than 2.0 cm (Table 3) and most Ki67 positive expression lengths were less than 3.0 cm (Table 3). The longest lengths of positive expression of p53 and Ki67 were 3.73 cm and the longest length of positive expression of Ki67 in the mucous layer of the distal end was 3.61 cm (Table 4).
Results of follow-up
Thirty four patients had died when the study closed. One patient had an anastomotic recurrence. Three-year survival was 48.4%. The main mode of treatment failure was lymph node and distant organ metastaseis.
DISCUSSION
The advantages and disadvantages of large pathologic sections
The process of making large pathologic sections is difficult, complex and time consuming. IHC staining of a large pathologic section is also more difficult and expensive than of a regular small section. Compared to the regular small sections, large pathologic sections have the advantage of offering a complete view of the esophagus, tumor and the relationship between the tumor and neighborhood tissue.[20] To our knowledge, our study is the first study that combined a large section technique and immunohistochemical analysis.
The shrinkage of esophageal specimens
Esophageal carcinoma specimens always shrink immediately after resection.[18,19] It is very important to measure the in situ length of the esophagus during the operation. The esophageal specimen undergoes further shrinkage during formalin fixation[18,19] and even during production of pathologic sections, there was a small amount shrinkage. We calculated the actual length during the operation by the principle of rational shrinkage to decrease the error due to the esophageal shrinkage.[20]
Current studies on the molecular resection margins
Molecular oncology has developed quickly in recent years. Several novel concepts have been introduced into the surgical oncology field, such as “molecular staging”, “molecular prognosis” and “molecular resection margin”. Brennan et al.[8] were the first to use the “molecular resection margin” concept. Many researchers performed similar studies in the head and neck cancer fields. Most of these studies used p53 as a molecular marker in examining the resection margins. [8-10] Some researchers have suggested the use of eIF4E as a molecular marker because the 4E proto-oncogene has been found to have almost 100% positive expression in head and neck cancers.[11,12] Some molecular margin research was also conducted on lung and esophageal cancers.[9,10] The advantage of using molecular resection margins is that we can detect occult malignant cells previously missed by physical examination and standard histopathologic techniques.
However, one of the limitations of studies published previously is that they only focused on the relationship between local recurrence and the positive molecular margin. Surgeons also need to know the state of the molecular maker expression beyond the margin. What is the optimal resection length to achieve negative molecular margins for esophageal squamous cell carcinoma? As far as we know, this study is the first study to demonstrate the extent of positive expression of molecular markers beyond the actual esophageal cancer.
Positive expression of the makers in the tumor
In our study, 66% of the patients with esophageal cancer displayed positive p53 expression, and 54% of the patients showed positive Ki67 expression. These tumor expression rates are similar to previous reports. 117,211 However, negative expression in the tumor for p53 was 34% and for Ki67 46%. This is a limitation for using p53 and Ki67 as molecular markers. eIF4E is a potential molecular marker, but we found no reports concerned with expression of eIF4E in esophageal cancer. In future studies, eIF4E should be tested.
What is the molecular resection margin when p53 is chosen as a molecular marker?
In a previous study of 23 patients, Shao et al.[9] reported that they used PCR to detect the p53 gene mutations in the resection margin tissue of esophageal squamous cell carcinomas. All the margins were negative by histopathological examination. There were 13 cases with p53 gene mutations in the resected margin tissue. The patients were followed for 3~16 months after the operation. Among the 13 cases with p53 gene mutations, 6 cases recurred. None recurred among the 10 cases without p53 gene mutations.
In our study of the resected mucous layers, 12 patients (17% of 70 cases) showed positive expression of p53 on the upper end and 14 patients (20% of 70 cases) displayed positive expression of p53 on the lower end. We found that these positive expressions usually were associated with either mild dysplasia or moderate dysplasia. These finding are similar to a previous report on p53 in esophageal cancer.[17]
The main purpose of our study was to determine the optimal resection length to achieve molecular margins free of esophageal squamous cell carcinoma. There was 83% 321 p53 positive mucosal expression within 2 cm from the tumor in the upper end and 86 within 2 cm from the tumor in lower end. The longest lengths of mucosal positive expression of p53 on the upper and lower ends were 3.33 cm and 3.73 cm respectively. We believe that esophageal tissue with positive p53 expression should be removed of because it has the potential to develop cancer. From our study, we concluded that at least a 4 cm margin on both upper and lower ends will leave esophageal tissue free of p53 expression.
What is the molecular resection margin when Ki67 is chosen as a molecular marker?
Ki67 usually is employed to measure the growth of normal tissues and malignant tumors.[16] Xu et al.[l7] reported that expression of Ki67 in normal, premalignant and malignant esophageal tissues was different. Our study is the first report using Ki67 as a molecular marker to analyze surgical margins.
In our study, positive expression of Ki67 in the mucous layer of the upper end was seen in 14 patients (20% of 70 cases) and in 16 patients (23% of 70 cases) of the lower end. We also found that positive expression was usually accompanied by mild dysplasia or moderate dysplasia. Findings are similar to a report on Ki67 in esophageal cancer.[17]
There was 57.1% Ki67 positive mucosal expression within 2 cm from the tumor in the upper end and 81.3% within 2 cm from the tumor in the lower end. The longest length of mucosal positive expression of Ki67 on the upper and lower end was 3.26 cm and 3.61 cm respectively. We believe that the esophageal tissue with the positive Ki67 expression should be removed to reduce tumor recurrence. We recommended that at least 4-cm margins should be obtained on both the upper and lower ends.
Pitfall of our study
One limitation of this study was that the positive expressions of p53 and Ki67 in the tumor were only 66% and 54%. We need to find some other molecular markers with a higher positive expression in the tumor. We examined only two molecular markers in our study. Other molecular tumor markers also need to be tested.
Summary
We used p53 and Ki67 as molecular tumor markers to study seventy specimens of esophageal squamous cell carcinoma on large pathologic sections. From our study, we found that both p53 and Ki67 showed positive expression beyond the tumor site, but remained within a distance of 4 cm. We conclude that at least 4-cm margins on both the upper and lower end are needed to achieve a molecular margin free of esophageal squamous cell carcinoma.
Footnotes
This work was supported by the China Guangdong Province Medical Science Research Grant (No. B2003053).
- Received September 12, 2006.
- Accepted October 16, 2006.
- Copyright © 2006 by Tianjin Medical University Cancer Institute & Hospital and Springer