Abstract
OBJECTIVE To investigate the frequency of BRCA1 gene mutations in breast cancer families in China.
METHODS Genomic DNA was obtained by conventional techniques from the peripheral blood mononuclear cells collected from 94 persons derived from 45 breast cancer families. All participants gave written informed consent. The mutations in the BRCA1 gene were detected by the polymerase chain reaction and single stranded conformation polymorphism (PCR-SSCP). Then, the samples of interest were sent for direct DNA sequencing.
RESULTS No mutation sites were found in exon 2 or 20 by DNA sequencing. Eight sites were found in exon 11 such as 2201C>T (Ser694Ser),3232A>G (Glu1038Gly),2201C >A/G (Ser694Arg),2731C>T (Pro871Leu),2086A>T (Asn591 lie) and three sites of 1584G>T (Glu424Stop). Three mutation sites were found in exon 16 which included 5106A >G (Met1663Val),5208delT (Stop1639) and 4956A>G (Ser1613Gly).
CONCLUSION These mutation sites may be related to breast cancer, but more investigation is needed to determine whether the mutation sites are hot spots of mutations in Chinese familial breast cancer patients.
keywords
The breast cancer susceptibility gene BRCA1 which is associated with hereditary susceptibility to breast and ovarian cancer is one of the first tumor-suppressor genes to be identified. Mutations in the BRCA1 gene are highly penetrant and confer a 85% risk of hereditary breast cancer.[1] There are some mutation sites that have been applied to screen the members of breast cancer families in Western countries in order to predict the risk of breast cancer.[2] In Asia, the BRCA1 mutation rates in breast cancer families have been published only from Japan, Singapore and Taiwan and have been reported to be lower than those of Western countries. Some reports from China are either from sporadic breast cancer patients or from limited breast cancer families, but there have been only a few reports concerning large scale BRCA1 mutation analysis of breast cancer families in China. In this study, we collected data from 45 breast cancer families selected from patients who visited or revisited the Tianjin Medical University Cancer Hospital during 1997~2003. The BRCA1 gene mutation analyseswere performed by the polymerase chain reaction and single conformational polymorphism (PCR-SSCP) and DNA sequencing in order to reveal the proportion of families who inherit the BRCA1 mutations, and the spectrum of the BRCA1 mutations in Chinese breast cancer families.
Materials and Methods
Subjects
Families were identified who were willing to participate in a genetic screening study of BRCA1 and who fulfilled any one of the following minimum criteria: there were more than 2 first degree relatives or second degree relatives with breast cancer in one family; there was one family member with ovarian cancer in addition to breast cancer; there was one male breast cancer patient in the family. A total of 45 breast cancer families were involved in our study, consisting of 94 participants, among whom 56 were breast cancer patients, and 38 were either first or second-degree relatives. Included were 2 male breast cancer patients. Control samples from a series of 39 normal individuals enrolled in one breast exam study were analyzed to determine population frequencies of observed variants.
DNA extraction
Genomic DNA was extracted from peripheral blood lymphocytes using a E.Z.N.A. Blood DNA kit.
Mutation analysis
Exon 2, 11, 16 and 20 of the BRCA1 gene were amplified by using 19 pairs of primers described in Table 1. PCR was carried out in a total volume of 50 μl containing 10 × buffer 5 μl, MgCl2 (25 mM)4 μl, dNTP (10 mM) 1 μ1, Taq DNA polymerase 1U (Takara), each primer (50 μM) 1 μl and genomic DNA 1-1.5 μg. The mixture was cycled at 94 °C for 30s, 50 °C for 50s, 72°C for a min, 35 times. Each 10 pl of the PCR products was diluted 1:1 in denaturing loading buffer (95% formamide, 0.05% bromophenol blue, 10 mM EDTA ), heated in boiling water for 10 min and cooled on ice for 10 min. Then a 20 μl sample was loaded on the gel (8%) for polyacrylamide gel electrophoresis (PAGE). The gel was run in 1 × TBE (Tris-Borate EDTA) under 100-Volts for 4~6 h at 4°C. After electrophoresis, the gels were silver-strained and the shift bands were examined.
The list of the sequence of primers applied to amplify the BRCAl gene
DNA sequencing
The shift bands were selected and the counterpart remaining PCR products were sent to the Takara Co. for completing bidirectional DNA sequencing.
Sequence analysis
DNA Star Mag-Align biology software was applied to compare our DNA sequences with the original DNA sequences of BRCA1 in order to find the mutation sites. Usage of BLAST 2 on the web of NCBI can reveal the alterations in proteins from the results of the DNA sequencing in order to ascertain whether the mutation sites can lead to amino acid changes (missense). Finally, the pathogenic mutations can be checked in the Breast Cancer Information Core database (Breast Cancer Information Core, http://www.nhgri.nih. gov/Intramural_research/Lab_transfer/Bic)so as to verify whether the mutations have been previously reported.
Results
PCR-SSCP results
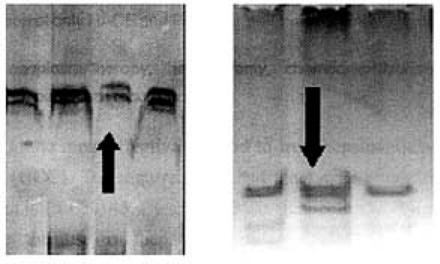
PCR products of all samples displayed single bands in PAGE compared to control samples, which indicated that the sequence of PCR products might have mutations (Fig. 1). In this study, 24 single shifted bands in the PAGE gel were found and were therefore sent for DNA sequencing(Table 2).
The shifted bands in PAGE.
The samples of doubted mutation sites
Analyses of the mutation sites (Table 3)
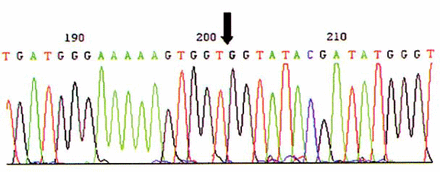
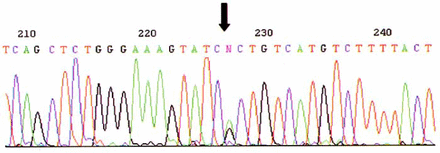
Eleven mutation sites were found by direct sequencing in 94 participants. No site was found in exon 2 or 20. Eight sites were found in exon 11 while 3 mutation sites appeared in exon 16. No.5 participant had a nucleotide substitution 3232A>G which leads to an amino acid change (GlulO38Gly)(Fig.2). No.17, 19 and 20 participants were three sisters from one family who harbored the same mutation site, 1584G>T (Glu424Stop), which resulted in a truncated protein. Another nucleotide change, 5208delT, produced a truncated protein in No.42. Two missense mutations were 5106A>G (Metl663Val) and 2731C>T (Pro871Leu), and one nonsense mutation was 2201C>T. A heterozygote mutation 2201C>A/G was found in No.32(Fig.3).
Polymorphism and sequence variants of the BRCA1 gene
The heterozygote mutation 3232A>G found in No.5.
The heterozygote mutation 2201C > A/G found in No.32.
Discussion
The BRCA1 gene is located on chromosome 17q21 and codes for a 7.5 kb transcript which spreads across 100 kb of genomic DNA. The gene consists of 24 exons among which 22 are coding. The first 10 and last 13 exons are relatively small whereas exon 11 represents over 60% of the entire coding sequence. The gene codes for a 1863 amino acid zinc finger containing protein of unknown function, which has been shown to interact with an additional protein termed BRCA1-associated ring domain protein (BARD1). [3] Recently, it has been reported that BRCA1-BARD1 heterodimers exhibit significant E3 Ub ligase activity and that the BARD1 RING finger domain greatly potentiates the ligase activity of the BRCA1 RING finger. This activity may play an important role in the regulation of the cell cycle, DNA repair in response to DNA damage and the inducement of tumor apoptosis.[4,5] Mutations in BRCA1 disrupt binding to BARD1, suggesting that interaction with BARD1 is significant in regulating BRCA1 function and the effect may even increase the risk of breast and ovarian cancer.[6]
Up to now, more than 300 mutation sites have been discovered in the BRCA1 gene. Some studies have shown that mutation hot spots of BRCA1 gene are dispersed in exons 2, 11, 16 and 20 in Western populations.[7] It is crucial to know whether there are mutational hot spots in inherited Chinese breast cancer families and if the incidence of breast cancer in China can be explained by the results from Western reports. In our study, no mutation sites were found in exon 2 and 20, which is consistent with the previous reports. [8,9] So we can draw the conclusion that the exons mentioned above were not the frequent regions of mutation in BRCA1. However, 8 mutation sites in the region of exon 11, one nonsense mutation and 7 missense mutations were found. The mutation, 3232A>G (Glul038Gly), which exists in the No.5 participant, is considered to be one hot mutation spot in the Chinese population for its high frequency has been reported. It was exciting that one mutation, 1584G>T(Glu489Stop), appeared in three sisters, one normal woman and two breast cancer patients. This mutation which leads to a truncated protein, has not been reported before and may be associated with the incidence of breast cancer in Chinese breast cancer families. We can infer that the sister without breast cancer is a high-risk person and should see a doctor for a breast examination at least two times a year. In this family, their mother was a breast cancer patient who died two years ago. It was regretful that a sample from their mother could not be obtained. The last mutation site in exon 11 was previously reported by a foreign researcher and has been considered to be a hot spot for a BRCA1 gene mutation in Western countries. The role of this mutation site has not been determined at present. The two mutation sites in exon 16 have not been found, so further studies for the identification of their mutation hot spot will be needed in the future.
The frequency of BRCA1 mutations in Chinese breast cancer families in our study is 10.6%, which is a little higher than that in the Chinese population reported previously and is lower than that of Western breast cancer families.[8, 10-14] The difference could be explained by their different mutation analysis methods, genetic background and environment. First, the methods of analyses included direct DNA sequencing, protein truncation tests (PTT), heteroduplex analysis (HDA), restriction fragment length polymorism (RFLP), DNA chips, denaturing high performance liquid chromatography ( DHPLC), single stranded conformational polymorphism (SSCP) and so on. PTT, HDA, RFLP and SSCP methods do not evince enough sensitivity for detection of mutations and as a result often produce false-negative results, while DNA chips and DHPLC need specific and expensive instruments. DNA sequencing is available and exact in Western countries but the price is costly in China. Most researchers in China perform multiple methods in order to make full use of limited research funds. Therefore, the results of testing for BRCA1 gene mutations in China are fewer than that in Western countries. Second, the subjects selected based on genetic background are too few or only those whose families have a close genetic relationship are enrolled as subjects. Third, apart from genetics, some carcinogenic factors such as hormonal levels, environmental factors and so on play a role in the etiology of breast cancer. It is probable that exposure to different carcinogenic factors in divergent areas results in a different incidence of breast cancer.
In our study, we have defined these BRCA1 gene mutations found in 45 breast cancer families. No other publications have reported as many breast cancer families in China as our report. It is very difficult to recruit a sufficient number of breast cancer families in China, as the lifetime risk of breast cancer for women in China is 4 or 5 times lower than that in the United States, and a registry for breast cancer has not been established in China. More work is needed to further clarify the relationship between the BRCA1 gene mutation and breast cancer pathogenesis.
- Received September 26, 2004.
- Accepted February 3, 2005.
- Copyright © 2005 by Tianjin Medical University Cancer Institute & Hospital and Springer