Abstract
Among central nervous system-associated malignancies, glioblastoma (GBM) is the most common and has the highest mortality rate. The high heterogeneity of GBM cell types and the complex tumor microenvironment frequently lead to tumor recurrence and sudden relapse in patients treated with temozolomide. In precision medicine, research on GBM treatment is increasingly focusing on molecular subtyping to precisely characterize the cellular and molecular heterogeneity, as well as the refractory nature of GBM toward therapy. Deep understanding of the different molecular expression patterns of GBM subtypes is critical. Researchers have recently proposed tetra fractional or tripartite methods for detecting GBM molecular subtypes. The various molecular subtypes of GBM show significant differences in gene expression patterns and biological behaviors. These subtypes also exhibit high plasticity in their regulatory pathways, oncogene expression, tumor microenvironment alterations, and differential responses to standard therapy. Herein, we summarize the current molecular typing scheme of GBM and the major molecular/genetic characteristics of each subtype. Furthermore, we review the mesenchymal transition mechanisms of GBM under various regulators.
keywords
Introduction
Gliomas are malignant primary tumors in the brain and/or spinal cord of the central nervous system (CNS); they have the highest rate of incidence and the poorest prognosis among CNS-associated malignancies. Among the clinical subtypes of glioma, World Health Organization (WHO) grade 4 glioma, also called glioblastoma (GBM), is the most lethal. In GBM, conventional treatment with surgery and chemotherapy (CT) leads to a 5-year survival rate of only 5.5%, possibly because of the carcinoma’s high molecular heterogeneity and chemoresistance, and the strongly invasive nature1. Notably, high inter- and intra-tumor heterogeneity are believed to cause chemo-resistance of GBM after long-term treatment with standard chemotherapeutics, thus resulting in poor patient prognosis and survival2,3. Attempts to classify the molecular characteristics of gliomas include subgroup classification schemes based on IDH, 1p/19q status, MGMT promoter methylation, or G-CIMP phenotype4,5. Other studies have reported GBM risk stratification by correlating the genome methylation status and corresponding transcriptomic changes, to improve the prognosis of patients with GBM6,7. However, none of these classifications are sufficient to characterize the molecular characteristics of GBM at the level of the whole gene expression profile. The studies of Phillips, Verhaak, and Wang et al. have proposed a GBM molecular subtyping scheme based on the expression profiles of GBM-associated genes; this scheme has been widely accepted for determining the molecular characteristics of GBM pathology8-10.
GBM phenotypes show elevated plasticity during and after CT, thus leading to a stage called proneural-mesenchymal transition (PMT), which is synonymous with the epithelial-mesenchymal transformation (EMT) in tumors of epithelial origin. We collectively refer to both PMT and EMT as mesenchymal transition (MT). Malignant phenotypes and chemo-resistant GBM have been associated with MT, and pose a substantial roadblock to the treatment response and tumor recurrence11,12. Because of differences in transcriptomic expression, tumor tissue characteristics, and immune cell infiltration levels across GBM phenotypes, sensitivity to CT and the prognosis also vary substantially among GBM subtypes13. The mesenchymal (MES) subtype is more aggressive than the proneuronal (PN) subtype and is strongly associated with poorer prognosis; even patients undergoing radiotherapy (RT) and/or CT can progress from PN to MES over the long term14.
Several clinical investigations have been conducted to understand the specific molecular characteristics and malignant behavior of the immune microenvironment, energy metabolism, tumor localization, and the presence of cancer stem cells in glioma. This review summarizes current research progress in exploration of the underlying pathomechanisms regulating the transition of a GBM subtype from early to malignant and recurrent phenotypes. The findings may provide avenues to novel CTs for preventing the recurrence of GBM tumors, thus providing more accurate targeted therapy strategies for patients with GBM.
Molecular classification of GBM
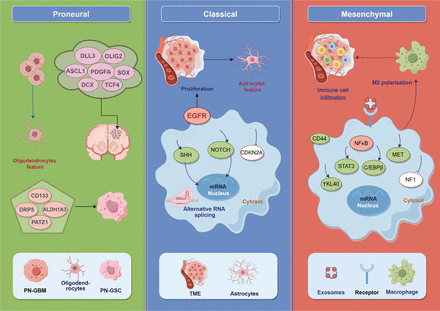
In 2006, Phillips et al. categorized high-grade gliomas (HGGs) into 3 subtypes: PN, MES, and proliferative (Prolif), according to the gene expression profiles of 76 astrocytoma tissues and 35 characteristic genes. Under this categorization, highly expressed PN and MES biomarkers are mutually exclusive, and the survival of the PN phenotype is significantly longer than that of the MES and Prolif phenotypes. For PN and MES subtyping, the expression profiles of DLL3 and YKL40 genes have high diagnostic and prognostic potential, independently of the tumor grade and tissue necrosis properties. Most HGGs follow one of these 3 subtype characteristics, and GBM with a dual subtype is clinically rare. Importantly, patients with HGG-PN tend to be younger than those with MES and/or Prolif subtypes. Patients with post-treatment relapsing HGG exhibit significantly elevated expression of MES markers, including YKL40, CD44, STAT3, and Vimentin (typical markers of mesenchymal tissue), and diminished expression of PN-associated OLIG2. Furthermore, the MES subtype shows elevated expression of angiogenic factors, including VEGF, VEGFR1/2, and PECAM1 (an endothelial marker). In the PN subgroup, loss of PTEN expression and significant activation of the Notch pathway are frequently detected, whereas the MES subgroup exhibits high AKT activation8.
In 2010, Verhaak and colleagues proposed 4 canonical subtypes of GBM—PN, neural (NE), classical (CL), and MES—on the basis of a cluster analysis of 200 gene expression profiles associated with GBM on 3 independent sequencing platforms. High EGFR expression was observed in 97% of CL subtype cases; EGFR expression and CDKN2A deletion were observed in 94% of cases, and significantly high expression of the Notch and Sonic hedgehog (SHH) pathway-linked genes was observed in the CL subtype. Oncogenic mutations in 6 of the 7 major tumor suppressor genes, including NF1 and PTEN, all of which are associated with AKT activation, have been reported in the MES subtype, thus adding to the previously identified upregulation of YKL40 and MET in that subtype. Interestingly, a combined expression profile of MES and the astrocyte markers CD44 and MERTK appears to represent active crosstalk between the MES subtype and EMT. Furthermore, TNF-α and NF-κB pathway-associated factors are highly expressed in the MES subtype. Three main features of the PN subtype are PDGFRA expression, mutations in IDH1 and TP53, and loss of heterozygosity. The PN subtype also exhibits high expression of oligodendrocyte maturation-associated genes such as PDGFRA, OLIG2, and NKX2-2, as well as pre-neurodevelopmental genes including DCX, SOX, ASCL1, DLL3, and TCF4. In contrast, the NE subtype shows elevated expression of neuronal markers such as NEFL, SYT1, GABRA1, and SLC12A5. On the basis of histological classification, the PN subtype is enriched in oligodendrocyte markers, whereas the MES subtype shows elevated expression of astrocyte and microglial markers. The CL subtype is closely associated with mouse astrocyte characteristics, and the NE type is associated with oligodendrocytes and astrocyte differentiation9.
In 2017, Wang et al. revisited deep molecular typing analysis of GBM subtypes. After exclusion of the mutant IDH1 phenotype, cluster analysis of the BFG/U133A collection reclassified the GBM with WT IDH1 into 3 subtypes: PN, CL, and MES10. All three isoforms have different characteristics from the NE isoform defined by Verhaak et al.9, thus suggesting that the NE subtype might not be tumor specific. The investigators of this study assumed that the NE subtype might have included contaminating normal tissue from the tumor periphery in the brain, thus further explaining why the NE subtype is the only subtype lacking the characteristic gene abnormalities. Wang et al. further observed lower tumor homogeneity in the MES subtype than the other subtypes. However, higher expression profiles of microglia, M1 and M2 macrophages, and glial cell infiltration markers were found in the MES subtype; this enhanced immune cell infiltration might be associated with NF1 gene deletion in this subtype. In addition, GBM has high cellular plasticity, such that GBM of the non-MES subtype tends to transform to the MES subtype after recurrence, and consequently exhibits enhanced immune cell infiltration, diminished tumor homogeneity, and elevated M2 macrophage infiltration scores. Notably, M2 macrophage infiltration plays an important role in the development of RT resistance in patients with GBM10.
The GBM tri-isotyping proposed by Wang et al. is now widely accepted because of its high consistency and stability. The tripartite scheme excludes the influence of IDH (mut) GBM10, in agreement with the most recent WHO classification of GBM15. Immunohistochemical (IHC) staining of subtype-specific marker genes (EGFR, PDGFRA, NF1, IDH1, P53, CD44, MERTK, and PTEN) can indicate the degree of malignancy in patients with glioma and aid in predicting their clinical prognosis. Likewise, simple IHC staining of GBM markers can help distinguish tumor subtypes in clinical samples and may facilitate analysis of large sample sizes16-18. Furthermore, 2 independent studies have reported the development of radiomics-based assays to better predict molecular subtypes and prognosis in patients with GBM19,20.
Molecular and clinical characteristics of GBM subtypes
PN-GBM subtype
Most non-GCIMP MES GBMs originate and evolve from PN-like precursor cells21. Likewise, glioblastoma stem cells (GSCs) reside on a single axis of variation, ranging from PN to MES22. The main marker genes of the PN subtype include DLL3, PDGFRA, OLIG2, SOX, DCX, ASCL1, and TCF4, which are also characteristic of oligodendrocytes and PN cells8-10. Although the prognosis of patients with PN subtype GBM is relatively good, Verhaak et al. have found that this subtype shows the least response to treatment. Importantly, Verhaak et al. do not believe that the subtype transition from PN to MES is part of the natural progression of GBM tumors9. Relatively higher brain volume in infiltrative peripheral edema may be associated with PN-GBM; therefore, the PN subtype might possess underlying vascular pathology23. PDGFA plays a crucial role in development of the PN subtype; moreover, the PN-GBM subtype can be modeled by induction of PDGFA expression in mice. In addition, PTEN downregulation facilitates tumorigenesis, and NF1 knockdown induces the transition of the PN subtype to the MES on a background of PDGFA knockdown21,24. Insulin receptor-insulin-like growth factor 1 receptor (IR-IGF1R) signaling plays an important role in the recurrence of GBM with high PDGF-PDGFR expression25. Moreover, expression of PATZ1 and DRP5 is specifically upregulated in PN-GSC and consequently promotes the maintenance of the PN-GSC stemness26,27. LGALS3 is an independent biomarker for patients with PN-GBM, who have relatively short overall survival (OS)28. Notably, Wee1 kinase is considered a potential therapeutic target in PN-GBM29. Moreover, Notch1 activation has been reported to promote the survival of GBM cells via the NF-κB pathway in CL-GBM and PN-GBM subtypes30.
CL-GBM subtype
The main features of the CL-GBM subtype are amplification of EGFR and loss of CDKN2A expression on the background of hyperactivation of Notch and SHH pathway-associated factors. In addition, the characteristic features of the CL-GBM subtype may be associated with those of mouse astrocytes9. Interestingly, although the PN and MES subtypes exhibit stemness properties, no solid evidence supports that the CL subtype also has cancer stemness features31. Most studies have focused on the 2 relatively mutually exclusive phenotypes between PN and MES subtypes, and have rarely investigated the CL subtype. The most likely reason is that the CL subtype does not possess the distinctive features of the other two subtypes. In one study, we first identified several candidate genes for different molecular subtypes, and then found that the radio- and chemo-resistant gene signatures were present only in the PN subtype described by Verhaak et al., but not in the PN subtype reported by Phillips et al. Next, we observed that the MES-GBM cells were under endoplasmic reticulum stress, and both angiogenesis and immuno-inflammatory responses were significantly elevated. Furthermore, we revealed that genes associated with alternative RNA splicing were significantly upregulated in the CL-GBM subtype, whereas the expression of genes associated with energy production was significantly elevated in the NE subtype32.
MES-GBM subtype
The major characteristics of the MES-GBM subtype include dominant NF1 gene mutation; NF-κB and AKT pathway activation; and elevated expression of YKL40, MET, and other signature markers in MES, angiogenic, and astrocyte cells. Significantly elevated heterogeneous and drug-resistant cancer cells, in addition to infiltrating immune cells, are also important hallmarks of this subtype8-10. Recent studies have shown significantly elevated endoplasmic reticulum stress in the MES subtype; moreover, MES-GBM might have similar characteristics to those of reactive astrocytes32,33. Whole transcriptome analysis of MES-GBM cells has revealed a characteristic competitive endogenous RNA (ceRNA) network that distinguishes the MES phenotype from other isotypes. Genes including RUNX1, TGFBR2, PPARG, GIT2, ACSL1, and RAP1B interact with protein factors in a ceRNA-regulated manner and have been identified as markers of the MES subtype34. In another study, C/EBPβ, C/EBPδ, FOSL2, STAT3, RUNX1, and bHLHE40 have been identified as transcription factors (TFs) in the MES subgroup35. Moreover, the RUNX1, FMNL1, and TGFBI genes have been reported to correlate with glioma staging and patient prognosis, respectively, and to serve as molecular markers for MES subtypes36-38.
Tumor microenvironment (TME)
The complexity of the TME is an important factor in GBM heterogeneity. Non-tumor cells, particularly brain-resident microglia and infiltrating monocyte-derived macrophages, are strongly associated with the MES-GBM phenotype39. Compared with the CL and PN subtypes, the MES subtype has high infiltration by non-tumor cells and shows high immune heterogeneity. Interestingly, although patients with MES-GBM show inflammatory and aggressive malignancy, they respond better to immunotherapy than patients with other subtypes10,40. IHC-based comparisons of characteristic immune factors have shown significant variation across GBM subtypes, with the MES subtype having the highest content of immune cells and the CL subtype appearing to be less conducive to immune cell infiltration41,42. The low OS of the MES subtype may be associated with its high immune and matrix infiltration43. Analysis of the regulatory transcription profiles of tumor-associated macrophages (TAMs) has suggested an association between TME components and MES-GBM pathogenesis44. Another study has indicated that RT might induce TAM infiltration into the TME, as observed in a pre-clinical GBM mouse model45. Furthermore, Dumas et al. have reported that GBM tumorigenesis is modulated by the mTOR-dependent STAT3 and NF-κB signaling pathways in microglia, thereby promoting an immunosuppressive microglial phenotype, particularly in the MES-GBM subgroup46.
Energy metabolism
In recent years, the metabolic differences uncovered among GBM subtypes have indicated that MES-GBM cells have significantly higher glycolytic activity than PN-GBM cells. Moreover, ALDH1A3 plays a key role in this context: significantly elevated expression of ALDH1A3 in MES-GBM cells enables rapid metabolism of glucose to lactic acid (LA), thereby stabilizing HIF-1α, activating the NF-κB signaling cascade, and inducing VEGF secretion by tumor-associated stromal cells—all of which are characteristics of MES-GBM. Inhibition of ALDH1A3 attenuates the MES subtype but not the growth of PN-GSCs47-49. Furthermore, MES-GBM cells exhibit a stronger necrotic phenotype than non-MES cells. Because LA accumulation typically occurs in and around necrotic areas, necrotic MES-GBM cells may serve as an additional source of LA molecules in the TME50. GBM metabolomics and transcriptome sequencing analyses by Heiland et al. have revealed that metabolites, including choline and LA, are closely associated with the immune and hypoxic microenvironments, which are strongly enriched in MES-GBM. That study has provided further support for the relevance of LA in MES characterization51. In addition, glutamine levels are significantly higher in MES-GBM cells than non-MES GBM cells; consequently, energy metabolism and tumor growth may be promoted by conversion of glutamate to pyruvate in mitochondria. Therefore, decreased glutamine levels suppress proliferation of the MES-GBM cell type52. Metabolomic analysis has indicated significantly increased triglycerides, phosphatidylcholine, and other types of phospholipids in the MES subtype. The proneural-like subtype is enriched in very long chain fatty acid lipids and glycerophospholipids with long-chain polyunsaturated fatty acids. Moreover, the PN subtype exhibits elevated creatinine and homocysteine levels but diminished levels of L-cysteine and palatinitol. In addition, the PN-like subtype shows higher expression of ACSL6 and phospholipids carrying docosahexaenoic acid than the MES-like subtype. Regarding docosahexaenoic acid metabolism, normal tissues have a phospholipid composition comparable to that in the PN subtype but markedly different from that in the MES subtype53. Transcriptomic and metabolomic studies of various glycorecombinant subgroups of GSCs have indicated abundant L-fucose and aberrant fucosylation activation in MES-GSC compared with PN-GSC. These features are present in both subtype-specific xenografts and patient samples, thus suggesting that L-fucose may be a potential GBM subtype-specific biomarker. In the MES-GBM pre-clinical model, pharmacological or genetic inhibition of fucosylation significantly decreases tumor growth. Mass spectrometry screening of glycans has indicated that the major fucosylated proteins are involved in extracellular matrix interaction, integrin-mediated signaling, and cell adhesion in treatment-associated GBM recurrence54.
Organizational positioning
Puchalski et al. have analyzed the anatomical transcriptional map of GBM through in situ hybridization, laser microdissection, and RNA sequencing, and have demonstrated that the heterogeneous transcriptional status of GBM cells in the TME can be explained by the anatomical heterogeneity in histology samples55. Local tissue biopsy and magnetic resonance imaging (MRI) findings, compared with the molecular expression profiles of GBM-associated factors from different anatomical sites of tumors, have indicated that the molecular typing depends primarily on the sample tissue localization, such that PN-GBM-linked genes are expressed primarily in the periphery, MES-associated genes are expressed in the center, and CL-associated genes are expressed in the middle of tumors. Moreover, the non-enhanced region in PN-GBM tumors is enriched in progenitor oligodendrocyte-specific proteins, whereas the non-enhanced region in MES-GBM tumors shows enrichment of astrocyte and microglia-specific proteins56,57. A study using laser microdissection to isolate RNA from the necrotic regions of GBM tumors has demonstrated that several important MES-associated factors, including TNF-α, HIF-1α, cell migration, and immune-related pathways, are enriched in perinecrotic or pseudopalisadation regions. Likewise, intensification of the necrotic process affects the transcription profile of GBM, such that the non-MES features become comparable to MES phenotypes34. However, more single-cell studies are warranted to determine whether these perinecrotic GBM cells have intrinsic MES signatures, given that these anatomical regions have been suggested to contain larger populations of TAMs than other tumor regions58.
Glioma stem cells
GSCs are a class of cells that maintain self-renewal, continue to proliferate, can propagate into multiple lineages, and can initiate tumorigenesis after secondary transplantation59. The heterogeneity of GBM-GSCs has been observed in recent studies60, and at least 2 distinct GSC subtypes, PN and MES, can be found in tumors61-63 distinctly expressing CD133 on PN-GBM cells and CD44 on MES-GBM cells64. Beyond differences in protein expression profiles, MES-GSCs show higher proliferation rates in vitro and in vivo than PN subtype cells after xenografting in the mouse brain. In addition, the MES-GSC phenotype is associated with a relatively shorter OS than other subtypes65,66. MES-GSCs are more angiogenic, invasive, and RT-resistant than the PN-GSC subtype48. However, whether the subtype specificity of GSCs is associated with common tumor cell subtypes remains to be demonstrated67. Compared with tumors of parental origin, GSCs differ in their transcriptomic and epigenetic profiles. Bhat et al. have assessed gene expression profiles of GSCs and xenografts in reference to their parental GBM profiles. Unexpectedly, most GSCs induced by the MES-GBM lost their MES signatures and exhibited higher expression of PN-GBM-associated genes. In contrast, all PN-GSCs maintained their PN status when examined in xenografts11. The main characteristics of the different GBM subtypes are shown in Figure 1.
Schematic diagram of the 3 classifications of GBM. GBM is classified into PN, CL, and MES subtypes, which express different characteristic genes. PN-GBM cells tend to have oligodendrocyte characteristics, CL-GBM cells have astrocyte characteristics, and MES-GBM subtypes show high immune cell infiltration (figure generated in Figdraw).
Mesenchymal transition regulators of GBM
Direct regulators
The phenotypic and genotypic plasticity of GBM cells is not only the major driver of intratumor heterogeneity but also a characteristic feature of the dynamic tumorigenesis process68. Cancer-associated primary transcriptional regulators are crucial for tumor growth and therefore may have therapeutic value69. The six major regulator networks of MES gene expression are ZNF238, STAT3, C/EBPβ, BHLH-B2β, FOSL2, and RUNX1, with C/EBPβ and STAT3 at the top of the network hierarchy order. Specifically, C/EBPβ and STAT3 are responsible for the reprogramming of neuronal stem cells to the MES subtype and conferring metastatic ability on glioma cells70. Transcriptional coactivators with a PDZ-binding motif (TAZ) trigger the expression of MES-associated factors in the PN-GSC in a transcription-enhancing association domain (TEAD)-dependent manner. Interestingly, the predicted TAZ downstream gene was not found to overlap with the target genes for STAT3 and C/EBPβ. Thus, TAZ might serve as an independent regulator of the MES characteristics and promote MT in GBM in multiple pathways71,72. NF-κB controls the expression of 3 important TFs, C/EBPβ, STAT3, and TAZ, thus enhancing MT in pre-glioma spheroid cultures, and leading to enrichment in CD44 subsets and radio-resistant phenotypes73,74. Furthermore, S100A4 is an upstream regulator of the primary EMT factors ZEB and Snail2, as well as other MT regulators in GBM75. The TF FOSL1 has been identified as a key regulator of the MES subtype, and its depletion can lead to loss of MES gene markers, and diminished stem cell characteristics and tumorigenic potential in vivo76.
Indirect regulators
Deubiquitinase and ubiquitin-binding enzymes play key roles in the regulation of cellular protein levels. Several studies have demonstrated that these enzymes promote MT in GBM. The deubiquitinases USP9X, USP21, and USP3 have been found to promote the MES transformation of GBM by stabilizing the expression of ALDH1A3, FOXD1, and Snail2, respectively, in the MES phenotype. These enzymes are also associated with GBM malignancy and poor prognosis. The USP9X inhibitor WP1130 and the USP21 inhibitor disulfiram have shown good therapeutic efficacy in MES-GSC-derived GBM xenograft models77-80. Furthermore, PRL1 promotes GBM progression by activating the USP36-mediated deubiquitination of Snail281. The ubiquitin-binding enzyme UBE2T regulates MT and GBM recurrence through the UBE2T/GRP78/EMT regulatory axis82.
FOXM1 is a member of the FOX TF family, and the FOXM1/ADAM17 feedback loop controls MT and regulates GBM progression via the EGFR/AKT/GSK3β signaling pathway83. The sialyltransferase ST3GAL1 regulates MT in GBM by targeting FOXM1 protein degradation via APC/C-Cdh184. GSCs with low CD90 expression show enhanced FOXS6 expression in glioma cells through IL-1 secretion, thereby activating the EMT pathway and promoting resistance to TMZ therapy in glioma cells85. Sortilin promotes GBM cell migration, invasion, and EMT through the GSK-3β/β-catenin signaling pathway. AF38469 (a novel sortilin inhibitor) may be a selective antineoplastic agent for sortilin overexpressing MES-GBM cells86,87. Nrf2 is overactivated in MES-GBM via a positive feedback loop between the SQSTM1/p62 and Nrf2 axes, thus promoting GBM proliferation, invasion, and MT88. RBPJ, a key TF in the Notch signaling pathway, promotes PMT in GBM by activating the IL-6/STAT3 pathway89. STAT3 directly modulates the expression of SLUG, thereby promoting MES conversion of GSCs90. DRR1 induces EMT activation through AKT phosphorylation, thus participating in GBM invasion and progression91. The IL-6 family cytokine OSM promotes both transcription and translation of YKL40 and fibronectin while decreasing expression of DLL3 and OLIG2 in GBM cells. OSM-regulated MES phenotypes are associated with enhanced MMP-9 activity, and cell migration, and invasion92. The PBX3-MEK-ERK1/2-LIN28-let-7b positive feedback loop promotes the MES phenotype, and PBX3 is a key modulator and potential therapeutic target for MT93. IMP1 induces activation of YAP/TAZ signaling by m6A modification, thereby facilitating MT of GSCs94. SRPX2 supports EMT via the MAPK signaling pathway, thereby promoting GBM transformation95. Finally, genes such as NKCC1, ESTRADIOL, and DUSP6 have been reported to play important roles in MT and developing invasive properties of GBM cells96-98.
NF-κB pathway-associated regulators
NF-κB is a ubiquitous TF with critical roles in the aggressive MES differentiation process in almost all types of cancers, including GBM. NF-κB is an upstream factor in the MES-GBM regulatory network and is known to promote MT in GBM by regulating STAT3, C/EBPβ, and TAZ expression74. Furthermore, NF-κB directly induces the expression of MES proteins (e.g., Twist1, CD44, N-cadherin, and Vimentin)99-101. The extracellular matrix glycoprotein tenascin C (TNC) mediates MT in GBM through NF-κB-dependent activation102. Hypoxia-induced hyperexpression of PLOD1 promotes the malignant phenotype of GBM through NF-κB signaling103. The chemokine CXCL1 is highly enriched in GBM and is positively correlated with poor patient prognosis. Elevated CXCL1 levels can confer RT resistance on GBM cells by activating NF-κB signaling, thus promoting the MT in GBM cells104. NUDT21 is an upstream regulator of the NF-κB pathway and a potential molecular target for MT in GBM105. ARPC1B expression supports the maintenance of MES-GBM phenotypic status as well as RT resistance by inhibiting TRIM21-mediated degradation of IFI16 and HuR, which activate NF-κB and STAT3 signaling, respectively, in GBM. AZD6738 inhibition by expression of ARPC1B in combination with RT has demonstrated excellent anti-GSC activity106. Chemerin-regulated paracrine and autocrine networks have been found to promote the MES features of GBM by inhibiting the ubiquitin-proteasome degradation of CMKLR1, a chemokine receptor expressed primarily on TAMs and partially on GBM cells. Elevated levels of CMKLR1 induce NF-κB pathway activation and MT107. The cell adhesion molecule CD146 stimulates cell survival signaling by inhibiting p53 expression and activating NF-κB by phosphorylation, thereby supporting GBM stemness properties, MT, and the development of radio resistance. CD146 has also been identified as an inducer of the oncogenic YAP108. The serine/threonine kinase MLK4 promotes MT in GBM by binding and phosphorylating NF-κB109. The high expression of GPX8 is associated with MES features and is negatively correlated with anterior PN features, thus suggesting that GPX8 may promote PMT in GBM. Correlation analysis has shown that GPX8 expression modulation is associated with the IL1-MYD88-IRAK-NF-κB pathway and immune infiltration in GBM110. Furthermore, the TGF-β-induced CLDN4/TNF-α/NF-κB signaling axis plays a key role in EMT in gliomas111. TGM2 is highly expressed in GBM necrotic regions, and, under regulation by the NF-κB signaling pathway, triggers MT in GBM-GSCs via modulating TFs including C/EBPβ, STAT3, and TAZ112.
Other independent regulators
LIF and CCL2 expression are strongly associated with malignant GBM intermediate subtypes and predict the survival of patients with GBM. In vitro expression of LIF and CCL2 induces the transcription of genes associated with MT in GBM cells113. Moreover, ALDH1A3, a major contributor to aldehyde activity, is a well-characterized driver of MT in GBM114. In addition, the NFAT2-HDAC1 pathway may also play important roles in maintaining malignant phenotypes and promoting the MT of GSCs, thus providing potential molecular targets for GBM therapy115. ME2 is positively correlated with MES-GBM characteristics, and it promotes the proliferation, migration, and invasion of glioma cells. ME2 induces the expression of MES markers, including MET, N-cadherin, YKL40, and Vimentin, while inhibiting expression of the PN gene OLIG2; consequently, ME2 may promote PMT in GBM116. In addition, CDCP1 is a transmembrane glycoprotein and a novel regulator of PMT in GBM117. The coding genes and their mechanisms of promoting MT are detailed in Table 1.
Coding genes of MT regulators
Non-coding RNAs
MicroRNAs (miRNAs)
miRNAs are non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. Some miRNAs modulate the expression of genes, such as E-CADHERIN, MMP9, MMP14, SLUG, MMP2, and SNAIL, that are associated with GBM invasion and PMT118. In contrast, miRNAs can inhibit MT by directly targeting and inhibiting the expression of factors such as the FZD6 and FZD7 receptors119,120; the transcription inhibitors ZEB1 and ZEB2121,122; proteins such as Smad2, LHFPL3, and ADAM19123–125; and factors associated with TGF-β126,127 and NF-κB128,129 pathways. Furthermore, some miRNAs are involved in cell-to-cell communication and intercellular interactions mediating MT in GBM130.
Long non-coding RNAs (lncRNAs)
lncRNAs, such as PDIA3P1, bind proteins, such as C/EBPβ, and consequently prevent ubiquitination-mediated degradation, thus acting as master regulators of the MT process. The small-molecule therapeutic NEF specifically targets p38α in brain cells and has excellent blood-brain barrier permeability. NEF at specific concentrations blocks upregulation of PDIA3P1 and confers synergistic synthetic lethality in TMZ therapy. The combination therapy of TMZ and NEF has demonstrated excellent synergistic antitumor effects both in vitro and in vivo131. LINC01057 interacts with IKKα, stabilizes its nuclear localization, and promotes chromatin accessibility at NF-κB responsive promoters through histone modification and NF-κB activation. IKKα knockdown disrupts the effect of LINC01057 overexpression on the PMT in GBM132. Our research group has shown that dysregulation of the lncRNA PRADX in GBM promotes MT by recruiting histone H3 trimethylated at lysine27 to chromatin through binding the EZH2 domain of the PRC2 complex and epigenetically regulating the STAT3 pathway133. Moreover, lncRNA regulates MT through ceRNA; downregulates the expression of TGF-β and NF-κB pathway-associated factors134,135; or exerts a sponging effect on Snail, and ZEB1, thereby promoting MT in GBM136,137. The molecular sponge mechanisms of lncRNAs also play roles in MT of GSCs138. Furthermore, some lncRNAs have been reported to participate in the MT in GBM cells by regulating the NF-κB and Wnt pathways; however, their specific regulatory mechanisms have not been explored139-141.
Radiotherapy
RT has been found to induce PMT in recurrent GBM144. Notably, NF-κB activation is a critical step in PMT initiation. Bhat et al. have demonstrated that the PN-GBM subtype can be transformed into the MES-like subtype after RT, as evidenced by the high expression of CD44 on the MES-GBM cell surface. Together, NF-κB activation and CD44 overexpression predict the treatment outcomes of CT and RT in GBM11. Additionally, STAT3 and C/EBPβ are key players mediating therapy-induced PMT. Thus, blocking STAT3 activation has been shown to suppress RT-induced PMT, thereby prolonging survival in PN-GBM mice. Ionizing radiation can transition GSCs initially enriched in CD133 PN characteristics to the CD109 expressing MES subtype in a C/EBPβ-dependent manner56,145. Furthermore, ionizing radiation treatment significantly increases PAK4 expression and nuclear localization. A novel nuclear PAK4/PPARγ complex is recruited to the Nox1 promoter, thereby upregulating Nox1 expression and activating MT in GBM cells146.
RT-treated GBM cells produce soluble intercellular adhesion molecule-1 (sICAM-1), which stimulates macrophage infiltration, thereby enriching the TME in inflammatory macrophages. As a paracrine factor, tumor-derived sICAM-1 activates macrophage secretion of WNT3A, which in turn promotes MT in GBM cells147.
Alternative splicing
Alternative splicing (AS) patterns are another form of intra-tumoral heterogeneity in GBM. Recent studies have shown that the GBM phenotype has a unique AS pattern148. Proteomic, transcriptomic, and functional analyses have confirmed that RSRP1 regulates AS in the MES-GBM subtype by modulating spliceosome assembly. RSRP1-regulated AS events result in skipping of PARP6 exon 18, thus forming a truncated oncogenic PARP6 variant that does not effectively inhibit NF-κB activation. Combined treatment of cultured GBM cells and GBM-tumor-bearing mice with spliceosomes and NF-κB inhibitors has shown synergistic effects on MES-GBM growth149.
Inflammatory factors and the immune microenvironment
Macrophages and microglia together constitute an integral part of the PMT in GSCs. RT-induced intratumor PMT is associated with NF-κB activation and macrophagic/microglial involvement in GBM11. The expression profiles of inflammatory genes are highly heterogeneous across molecular subtypes of GBM, and are enriched in the MES subtype but moderately downregulated in the PN phenotype. Other inflammation-associated processes, such as the TAM signature, are upregulated in the MES subtype and result in poor prognosis. We have found that several GBM tumor-associated proteins, such as IL-6, IL-8, and CCL2, are actively expressed in GBM cell lines and have differential and synergistic roles in promoting proliferation, angiogenesis, invasion, and macrophage polarization in vitro150. The inflammatory chemokine CXCR4 is a predictor of poor prognosis in patients with GBM and can be used as a biomarker for the MES-GBM subtype. Moreover, CXCR4 mediates MAPK signaling pathway activation, specifically in patients with MES-GBM151. CXCR4 signaling promotes PMT and shortens the OS in patients with GBM, thus suggesting its inhibitory role as a potential therapeutic strategy152.
Abnormal vascularization
Resistance to anti-angiogenic therapy is associated with MT in patients with GBM153. Compared with the PN and CL subtypes, the MES subtype expresses higher levels of angiogenic markers8,35. A robust transformation of vascular endothelial cells into MES-GSC-like cells in GBM has been reported to induce chemoresistance through Wnt-β-catenin signaling activation154. Another study has identified that the PDGF/NF-κB/Snail axis induces MT by decreasing VEGFR2 expression in ECs. The dual inhibition of VEGFR and PDGFR eliminates tumor-associated ECs and prolongs OS in mice with GBM. Collectively, these findings shed light on the plasticity of ECs in controlling resistance to anti-angiogenic therapy, and suggest that vascular deconversion may be a promising treatment option for anti-angiogenic therapy155.
Exosomes
The molecular subtypes of GBM and GSCs differ in the expression of extracellular vesicle (EV)-associated genes, and GSCs with the PN or MES subtype, thereby producing EVs with significantly different characteristics, marker profiles, proteomics, and endothelium stimulating activities61.
GBM and EC-secreted exosomes have been shown to promote PMT in GBM and GSC cells by activating NF-κB and STAT3 pathways, thus upregulating MMP expression, and inhibiting the Notch pathway156,157. These findings further suggest that macrophage-derived EVs may be key regulators of PMT in GSCs by delivering miRNAs and promoting the MT of GSCs by targeting CHD7158.
Hypoxia
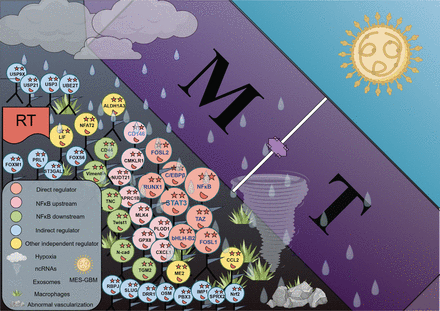
Hypoxic cells activate pro-angiogenic factors, including VEGF/VEGFR, PDGFR, and TGF-β, by stabilizing HIF-1/2α and consequently inducing transcriptional changes leading to the recruitment of inflammatory cells and facilitating PMT159. Joseph et al. have revealed that hypoxia aggravates the invasive nature of GBM cells by promoting ZEB1-HIF1α axis-mediated MT160. EPHB2 is epigenetically overexpressed in hypoxia, a common condition in malignant tumors. In addition, HIF-2α is required to stabilize EPHB2 during hypoxic shock. Finally, we have found that overexpression of EPHB2 promotes GBM invasion through phosphorylation of paxillin in hypoxia. These findings establish the HIF2α-EPHB2-paxillin axis as a regulatory mechanism for EMT161. Other MT regulators and their mechanisms are shown in Table 2. The schematic diagram of the occurrence of MT in GBM under the influence of various regulators is shown in Figure 2.
Schematic diagram of MT in GBM. Under the influence of many regulators represented by NF-κB, as well as many microenvironmental factors such as hypoxia, exosomes, and macrophages, GBM cells undergo mesenchymal transformation (figure generated in Figdraw).
Other mesenchymal transition regulators
Conclusions
The GBM molecular subtype is not a stable phenotype but a dynamic state with spontaneous changes in the TME74. Many studies have examined the PN and MES subtypes; however, data on the CL subtype are scarce. Importantly, the CL-GBM subtype accounts for a considerable proportion of GBM that has high EGFR expression and undergoes rapid proliferation, thus leading to poor prognosis; this subtype resembles the Prolif-GBM subtype identified by Phillips et al8. Because factors including hypoxia, angiogenesis, RT, and the immune microenvironment are associated with the MES phenotype, specific inhibitors of the MES phenotype in combination with other regimens should be considered to synergistically inhibit GBM progression and recurrence. However, caution is warranted, because polypharmacy may be associated with therapeutic complications. For example, the combination of TMZ with bevacizumab results in high toxicity and intracranial hemorrhage in patients with GBM162.
IHC detection of expression of GBM-specific genes is currently routinely used in clinical practice for patients with GBM, and the WHO 2016 classification of CNS tumors is used as a reference beyond histology to define the staging of solid tumors. This approach proposes a concept of how to construct a diagnostic model for CNS-specific tumors in the molecular era163. The most recent WHO classification, the 2021 edition, further emphasizes the significance of the determination of molecular characteristics of GBM subtypes, tissue typing and treatment regimen selection, and histological classification. However, the specificity of these histological subgroups based on the expression profiles of single or multiple factors is not sufficient to fully represent the complexity of GBM histology at the molecular level. Therefore, future research is required to establish clearer histological and molecular parameters15. The stability of the GBM molecular typing scheme has been fully validated by Wang et al.10; therefore, we believe that the molecular typing of GBM has great potential in guiding GBM diagnosis and therapy in clinical practice.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Chuan Fang, Yanli Tan, Can Xu.
Collected the data: Xiang Li, Menglin Xiao, Ziqi Zhang, Ziru Li.
Contributed data or analysis tools: Jianglong Xu, Guoming Liu.
Performed the analysis: Pengyu Hou, Xiang Li.
Wrote the paper: Can Xu, Pengyu Hou.
Footnotes
↵*These authors contributed equally to this work.
- Received December 27, 2023.
- Accepted March 28, 2024.
- Copyright: © 2024, The Authors
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.
- 79.
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵
- 88.↵
- 89.↵
- 90.↵
- 91.↵
- 92.↵
- 93.↵
- 94.↵
- 95.↵
- 96.↵
- 97.
- 98.↵
- 99.↵
- 100.
- 101.↵
- 102.↵
- 103.↵
- 104.↵
- 105.↵
- 106.↵
- 107.↵
- 108.↵
- 109.↵
- 110.↵
- 111.↵
- 112.↵
- 113.↵
- 114.↵
- 115.↵
- 116.↵
- 117.↵
- 118.↵
- 119.↵
- 120.↵
- 121.↵
- 122.↵
- 123.↵
- 124.
- 125.↵
- 126.↵
- 127.↵
- 128.↵
- 129.↵
- 130.↵
- 131.↵
- 132.↵
- 133.↵
- 134.↵
- 135.↵
- 136.↵
- 137.↵
- 138.↵
- 139.↵
- 140.
- 141.↵
- 142.↵
- 143.↵
- 144.↵
- 145.↵
- 146.↵
- 147.↵
- 148.↵
- 149.↵
- 150.↵
- 151.↵
- 152.↵
- 153.↵
- 154.↵
- 155.↵
- 156.↵
- 157.↵
- 158.↵
- 159.↵
- 160.↵
- 161.↵
- 162.↵
- 163.↵