Abstract
Objective: Leptin (LEP) is an obesity-associated adipokine associated with tumor cell growth. We examined the relevance of genetic variants of LEP and leptin receptor (LEPR) to colorectal cancer (CRC) survival by using data from the Newfoundland Familial Colorectal Cancer Study.
Methods: A total of 532 patients newly diagnosed with CRC between 1997 and 2003 were followed up until April 2010. Data on their demographics and lifestyles were collected via questionnaires. Genotyping of blood samples was performed with the Illumina Human Omni-Quad Bead chip. Multivariable Cox models were used to assess the relationships of 35 tag single-nucleotide polymorphisms (SNPs) in LEP and LEPR with overall survival (OS), disease-free survival (DFS), and CRC-specific survival.
Results: At the gene level, LEP was associated with DFS (P = 0.017), and LEPR was associated with both DFS (P = 0.021) and CRC-specific survival (P = 0.013) in patients with CRC. In single-SNP analysis, LEP rs11763517, LEPR rs9436301, and LEPR rs7602 were associated with DFS after adjustment for multiple testing. The LEPR haplotypes G-C-T (rs7534511-rs9436301-rs1887285) and A-A-G (rs7602-rs970467-rs9436748) were associated with prolonged OS among patients with CRC overall (G-C-T: HR, 0.63; 95% CI, 0.43–0.93; A-A-G: HR, 0.59; 95% CI, 0.38–0.91) and those diagnosed with colon cancer (G-C-T: HR, 0.54; 95% CI, 0.34–0.86; A-A-G: HR, 0.49; 95% CI, 0.29–0.83). Similar results were observed for DFS. Moreover, significant interactions were found among LEPR rs7602 (A vs. G), LEPR rs1171278 (T vs. C), red meat intake, and BMI status: the associations between these variants and prolonged DFS were limited to patients with below-median red meat consumption and body mass index (BMI) < 25 kg/m2.
Conclusions: Polymorphic variations in the LEP and LEPR genes were associated with survival of patients after CRC diagnosis. The LEP/LEPR-CRC survival association was modified by participants’ red meat intake and BMI.
keywords
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the second most deadly cancer worldwide1. Obesity is a complex epidemic disease involving metabolic alterations that can trigger various other diseases, including CRC. The mechanisms through which obesity is associated with increased CRC risk may be explained by factors such as hyperinsulinemia, oxidative stress, inflammation, and alterations in adipokine concentrations2. Leptin (LEP), the most abundant adipokine, has key roles in suppressing appetite and food intake, thus regulating energy homeostasis and body weight3. High levels of serum LEP have been detected in obese people, who have LEP resistance and thus do not benefit from LEP’s anorexigenic effects4.
LEP has been associated with elevated risk of developing CRC, as supported by in vitro, in vivo, and large epidemiological studies5. By binding its receptor, LEPR, LEP stimulates cell proliferation, inhibits apoptosis, and promotes angioneogenesis at various levels via several signaling pathways (e.g., JAK2/STAT3, PI3K/AKT, and MAPK/ERK)5–7. LEP also has proinflammatory properties that promote colon carcinogenesis5.
Polymorphisms in the LEP and LEPR genes are increasingly being studied in conjunction with LEP levels, to provide insight into their roles in obesity-mediated cancers, although data directly linking LEP and LEPR genetic variations to CRC are limited. Several single-nucleotide polymorphisms (SNPs) have been implicated in CRC pathogenesis; notably, the LEP rs2167270 (GG) and LEPR rs12037879 (GA/GG) genotypes are associated with elevated risk of CRC8–11. LEP and LEPR have also been suggested to be involved in survival after CRC diagnosis. Research has suggested that LEP mRNA expression levels are upregulated in colon cancer tissue and are associated with poor prognosis in patients with colon cancer12,13. Similarly, LEPR is overexpressed in primary CRC relative to normal colonic mucosa; intriguingly, however, LEPR positive tumors have been associated with superior overall survival (OS) in patients14,15. Nevertheless, no study to date has examined the polymorphic profiles of the LEP and LEPR genes in relation to CRC survival. Current understanding of the link between polymorphic variants and CRC survival is based on contradictory and inconclusive data suggesting a potential association of LEP/LEPR genetics with cancer risk.
Beyond the inherited genetic background, environmental components and their interactions interfere with CRC initiation and progression. However, the relationships of LEP and LEPR with modifiable lifestyle factors [e.g., intake of red meat and body mass index (BMI)], particularly the extent to which lifestyle factors may modulate this genetic risk, remains unknown. Thus, potential gene-environment interactions require further investigation to provide new insights that may lead to novel therapeutic targets and prevention strategies.
We therefore analyzed genetic variation in the LEP and LEPR genes in relation to CRC survival through a tag SNP approach to probe common genetic variations and construct haplotype blocks in the 2 genes. We further examined whether these associations might be modified by behavioral risk factors.
Materials and methods
Study population
The data were drawn from the Newfoundland and Ontario Familial Colorectal Cancer Study, a population-based cohort study investigating environmental and genetic components in CRC. The study methods and detailed rationale have been described before16–18. In brief, the participants were newly diagnosed with CRC between 1997 and 2003 and were 20–75 years of age at the time of diagnosis. A total of 532 patients with CRC (202 women and 330 men) residing in the provinces of Newfoundland and Labrador were identified through the Newfoundland Familial Colorectal Cancer Registry. The Health Research Ethics Authority of Memorial University of Newfoundland approved the study (Approval No. 40001511). Informed consent was obtained from each patient before participation.
Diet assessment and baseline information collection
At baseline, all patients with CRC completed a detailed family history questionnaire, a personal History Questionnaire, and a food frequency questionnaire, in which information on demographics, lifestyles, and dietary habits was gathered. All questionnaires were self-administered with a reference period of 1 year before diagnosis, to capture pre-diagnosis information. The median time from the date of diagnosis to questionnaire completion was 1.8 years. The dietary questionnaire was adapted from the Hawaii semi-quantitative food frequency questionnaire and was validated in the Newfoundland population19. BMI was defined as the weight in kilograms divided by the square of the height in meters. Information on weight and height was self-reported, and obtained from the PHQ with the following questions: “About how tall are you, without your shoes on?” and “How much did you weigh about 1 year before your recent cancer diagnosis?” Self-reported measures of weight and height are believed to be valid alternatives for determining weight status20.
Study outcomes
All study participants were followed, and death, cancer recurrence, and metastasis from the date of diagnosis until April 2010 were recorded. The endpoints for this study included OS, defined as the time from CRC diagnosis to death due to any cause; disease-free survival (DFS), defined as the time from CRC diagnosis to death due to any cause, CRC recurrence, or metastasis, whichever came first; and CRC-specific survival, measured from the date of diagnosis to the date of death due to CRC.
SNP selection and genotyping
Genotyping of peripheral blood samples from participants was performed at Centrillion Biosciences (USA) with the Illumina Human Omni-Quad Bead chip, which contains approximately 1.1 million SNPs. Additionally, 200 duplicates were genotyped with the Affymetrix Axiom myDesign GW Array Plate, which contains 1.3 million probes. SNPs with genotype concordance < 97% between platforms were excluded from this analysis.
Data cleaning and quality control filtering were conducted with Plink v1.07. Tag SNPs capturing common genetic variations in the candidate genes were selected with Plink v1.07 according to the following criteria: minor allele frequency > 5%; HWE P > 0.001; and linkage disequilibrium (LD) pruning with the Plink option “-indep-pairwise 50 2 0.8.” This process identified 3 SNPs for LEP and 29 SNPs for LEPR. We additionally included 3 high interest SNPs reported in previous studies (LEPR rs1137101, LEPR rs1137100, and LEPR rs1805096).
Specific information on MSI testing and mutation detection on BRAF V600E in tumor DNA has been reported previously18. MSI status in CRCs was determined by DNA testing with 5–10 microsatellite markers. An allele-specific polymerase chain reaction technique was used to detect mutant alleles in the BRAF gene18.
Statistical analysis
The log rank test was used to compare the survival distributions of the baseline characteristics. We tested the overall association of genes with principal component (PC) analysis. Single-SNP analysis and haplotype analysis were used to further explore variants in the LEP and LEPR genes in relation to CRC survival. The PCs were modeled with Cox proportional hazards regression, by using at least an 80% explained-variance threshold for determining the number of PCs to retain in the models. With the likelihood ratio test, we calculated the P-values for global associations between genes and disease outcomes by comparing 2 models with and without selected PCs, with degrees of freedom equal to the number of PCs. All analyses were stratified by anatomical site (i.e., the colon and rectum).
The data were further explored in a single-SNP analysis, with an additive model by Cox regression analysis. The decision for variable inclusion in the final model was based on statistical significance, according to stepwise regression with a P-value threshold of 0.05. The covariates eventually included in the model were age at diagnosis, gender, race, stage at diagnosis, household income, reported screening procedure, marital status, family history, smoking status, alcohol consumption status, folate intake, MSI status, and BRAF mutation status. Hazard ratios (HRs) and 95% confidence intervals (CIs) were applied to estimate the relationships of individual SNPs with OS, DFS, and CRC-specific survival. To control for type I error inflation, a multiple comparison adjustment specifically created for correlated tests due to LD was used21.
LD plots were generated with Haploview version 4.2 to identify haplotype blocks. PHASE version 2.1 was used to estimate the haplotypes in each block. The relationship between haplotype and survival in patients with CRC was assessed with Cox regression modeling, with reference to the most common haplotype. Bonferroni correction for multiple testing was performed for 37 haplotypes, thus yielding an adjusted P-value of 0.0014. A global P value for each haplotype block was obtained with a likelihood ratio test. Gene-environment interactions were estimated with stratified analyses (by intake of red meat and BMI) and the Wald method through introduction of a multiplicative interaction term into the model and assessment of its significance. Analyses were performed in SAS software version 9.4 (SAS Institute, Cary, NC, USA) and GraphPad 8.0.2. All tests were 2-sided.
Results
Patient characteristics and clinical predictors
The study population consisted of 330 men and 202 women. The mean age of the study population was 60.1 ± 9.2 years; 72.4% of the participants had a history of smoking; 96.9% of the participants were white; and 11.5% reported a bowel screening history (Table 1). Information on MSI status was obtained in 504 patients, of whom 11.5% were classified as MSI-H, and 88.5% were classified as MSS/MSI-L. In this study, almost all (96%) patients received surgery, and 21% underwent radiation or chemotherapy. At the end of the follow-up, 183 of the 532 patients had died. Most deaths (90.4%) were due to CRC. In the log-rank univariate analysis, male, advanced stage at diagnosis (IV), non-white race, chemoradiotherapy, consumption of > 3 servings of red meat per week, and MSS/MSI-L tumors were significantly associated with shorter OS, whereas bowel screening procedure, smoking status, alcohol consumption status, surgery, and BRAF mutation status were not associated with OS.
Demographical and clinicopathological characteristics of patients in the Newfoundland Familial Colorectal Cancer Study
Association of LEP and LEPR with survival in patients with CRC
To evaluate the overall gene-level association between LEP or LEPR and CRC survival, we performed PC analysis with OS, DFS, and CRC-specific survival as the endpoints (Table 2). At the gene level, LEP was significantly associated with DFS (LEP, global P = 0.017), and LEPR was associated with both DFS (global P = 0.021) and CRC-specific survival (global P = 0.013), both overall and stratified by colorectal subsite. However, we did not observe any meaningful relationship between LEP or LEPR and OS.
Associations of LEP and LEPR genes with colorectal cancer overall survival, disease-free survival (n = 532), and CRC-specific survival (n = 459)
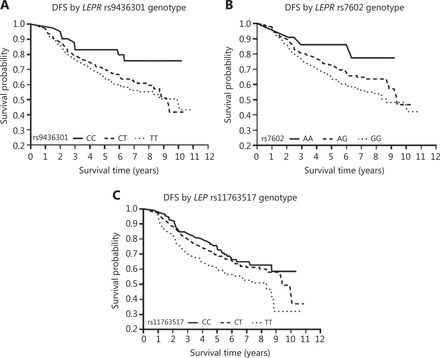
The relationships between individual SNPs within each gene and CRC survival were then evaluated with additive models (Supplementary Tables S1 and S2). Evaluation of 3 SNPs in LEP and 32 SNP in LEPR revealed 2 SNPs significantly associated with OS, 6 SNPs significantly associated with DFS, and 8 SNPs associated with CRC-specific survival. However, after adjustment for multiple testing, only LEP rs11763517 (Punadjusted = 0.001; Padjusted = 0.015), LEPR rs9436301 (Punadjusted = 0.000; Padjusted = 0.010), and LEPR rs7602 (Punadjusted = 0.000; Padjusted = 0.008) were associated with the DFS of CRC. Specifically, for LEP rs11763517, the C-allele was associated with longer DFS than the T-allele (HR, 0.64; 95% CI, 0.50–0.83) (Figure 1); a similar protective effect of the C allele on DFS was found for LEPR rs9436301 (HR, 0.60; 95% CI, 0.45–0.79). For LEPR rs7602, the A-allele was associated with longer DFS than the G-allele (HR, 0.55; 95% CI, 0.39–0.76). No significant relationship was observed between any SNPs in LEP or LEPR and overall or CRC-specific survival, after adjustment for multiple comparisons. When we repeated the analyses in patients with CRC diagnosed at a later stage (i.e., stage III/IV), we observed similar results for DFS (LEP rs11763517, HR, 0.63; 95% CI, 0.44–0.91; LEPR rs7602, HR, 0.39; 95% CI, 0.24–0.63; and LEPR rs9436301, HR, 0.55; 95% CI, 0.37–0.82). In addition, this association was more pronounced in advanced-stage cancers (stage III/IV) (LEPR rs7602 HR, 0.39, 95% CI, 0.24–0.63; LEPR rs1171278, HR, 0.40, 95% CI, 0.24–0.68) than in those detected at an early stage (stage I/II) (LEPR rs7602, HR, 0.82, 95% CI, 0.51–1.31; LEPR rs1171278, HR, 0.83, 95% CI, 0.49–1.40) (Supplementary Tables S3 and S4). Furthermore, analysis stratified by MSI status indicated similar patterns of association for MSI-H and MSS/MSI-L tumors (data not shown).
(A) Disease-free survival curves for patients with the LEPR rs9436301 genotype. (B) Disease-free survival curves for patients with the LEPR rs7602 genotype. (C) Disease-free survival curves for patients with the LEP rs11763517 genotype.
Haplotypes and survival of patients with CRC
Haplotype analysis assessed 3 SNP sites of LEP and 32 SNP sites of LEPR for LD (Supplementary Figures S1 and S2). We identified a total of 10 haplotype blocks of LEP and LEPR, and ignored combinations with frequencies less than 0.01 in the analysis (Table 3). For the LERP gene, the haplotype G-C-T in LD block 2 (rs7534511-rs9436301-rs1887285) was associated with longer OS among patients with CRC overall (HR, 0.63; 95% CI, 0.43–0.93) and patients with colon cancer (HR, 0.54; 95% CI, 0.34–0.86) than the most common haplotype. The results were similar for DFS (CRC, HR, 0.55; 95% CI, 0.38–0.80; colon cancer, HR, 0.54; 95% CI, 0.34–0.86). The haplotype A-A-G situated in LD block 3 of LEPR (rs7602-rs970467-rs9436748) was significantly associated with increased OS (HR, 0.49; 95% CI, 0.29–0.83) and DFS (HR, 0.58; 95% CI, 0.34–0.99) for patients with colon cancer. Notably, SNP LEPR rs7602, which had been previously identified as being inversely associated with mortality risk in single SNP analysis, was embedded within this haplotype. However, these associations were rendered nonsignificant after adjustment for multiple testing.
Associations of LEP and LEPR gene haplotypes with overall survival, disease-free survival (n = 532), and CRC-specific survival (n = 459)
Gene-environment interactions
To evaluate potential gene-environment interactions, we cross-tabulated red meat consumption, BMI, and LEP and LEPR polymorphisms among the participants (Table 4). Superior DFS was associated with LEPR rs7602 (A allele vs. G allele: HR, 0.48; 95% CI, 0.28–0.81) in the stratum of patients with red meat intake below the median. Additionally, LEPR rs1171278 (T allele vs. C allele: HR, 0.24; 95% CI, 0.09–0.62) was associated with greater prognostic benefits in patients with a BMI below than above 25 kg/m2.
Associations between selected genetic variations in LEP or LEPR and colorectal cancer disease-free survival, stratified by red meat intake and BMI
Discussion
In this study, the LEP and LEPR genes were associated with DFS and CRC-specific survival in CRC, respectively, at the gene level. Notably, LEP rs11763517, LEPR rs9436301, and LEPR rs7602 polymorphisms exhibited significant associations with DFS in patients with CRC after adjustment for multiple comparisons. Haplotype analyses indicated that the LEPR block 2 haplotype G-C-T, defined by rs7534511, rs9436301, and rs1887285, and the block 3 haplotype A-A-G, defined by rs7602, rs970467, and rs9436748, were associated with prolonged OS and DFS among patients with CRC and colon cancer. Furthermore, the LEP/LEPR-CRC survival association appeared to be modified by red meat intake and BMI.
LEP, which acts as a growth factor in colonic epithelial cells, might underlie the observed associations among obesity, physical activity, and colon cancer22. Various polymorphisms in the LEP gene are associated with extreme obesity (BMI ≥ 40 kg/m2)23, and LEP concentration is positively correlated with BMI24. Leptin is an important adipokine believed to play critical roles in stimulating proliferation and inhibit apoptosis25. Previous studies have shown that LEP up-regulates miR-4443, thereby suppressing NCOA1 and TRAF4, and decreasing the invasiveness of human colon cancer cells26. Chronic increases in LEP concentration may enhance the growth of colonic cancers via the MAPK and PI3-K pathways27. LEP has been implicated in breast cancer, prostate cancer, and diffuse large B-cell lymphoma28–30. LEP and LEPR gene polymorphisms are associated with the risk of CRC31,32, but no studies have described LEP or LEPR polymorphisms and CRC survival. In the current study, we identified SNPs in the human LEP and LEPR genes associated with CRC survival, which have not been reported in previous studies. Specifically, rs11763517, which is located in the intron of LEP (hg19: chr7:127890062), was associated with prolonged DFS. Potential mechanisms underlying this observation may be that LEP influences the growth and survival of CRC stem cells, and regulates the adhesion and invasion of colorectal carcinoma cells through activation of the JAK and ERK signaling pathways33. However, the association between the LEP rs11763517 genotype and LEP expression remains to be determined. In-depth understanding of the mechanism between LEP rs11763517 and CRC survival will require further investigation.
The physiological mechanisms of LEP action are exerted through LEPR, which is often expressed in CRC. A cohort study has indicated that elevated LEPR expression is associated with increased neoangiogenesis and metastatic potential in CRC34; and the absence of LEPR expression is correlated with low rates of proliferation35. The rs7602 variant is in the intron of LEPR (hg19: chr1:65897951) and the 3′UTR of LEPR overlapping transcript (LEPROT); the protein encoded by LEPROT has been shown to negatively regulate LEPR expression in mice36. Kim et al.37 have demonstrated that LEPR rs7602 is significantly associated with the risk of late menarche associated with decreased CRC risk and all-cause mortality38–40. However, results from the NIH-AARP Diet and Health Study have not indicated the same association between reproductive/hormonal factors and CRC41. LEPR rs9436301 is a variant in the second intron of LEPR (hg19: chr1:65895927) that has been reported to be independently associated with LEPR expression levels42. Studies have shown that SNPs within introns have the potential to affect the alternative splicing of RNA43, and 3′UTR variants are strongly associated with human traits and diseases44. In the current study, none of the high-interest SNPs in the literature (rs1137101, rs1137100) were significantly associated with CRC survival. Haplotype analysis indicated that the haplotype G-C-T in block 2 and haplotype A-A-G in block 3 on LEPR were associated with prolonged OS and DFS among patients with CRC overall and patients with colon cancer, as compared with the most common haplotype. These 2 haplotypes contained SNPs that were significantly associated with CRC survival in the single SNP analysis. Notably, haplotype analysis provides insights into genetic diversity and thus may be superior to individual SNP analysis.
Furthermore, our study is the first to demonstrate that red meat intake and BMI status may modulate the relationships between LEPR rs7602 or LEPR rs1171278 and CRC survival. Although the mechanisms underlying the observed interactions are not yet fully understood, the positive associations among red meat intake, BMI, and LEP levels, as demonstrated in previous research45,46, may provide an explanation. In addition, we surmised that the influence of subtle differences among genotypes was overwhelmed by the detrimental effects of overweight/obesity or high red meat consumption on cancer outcomes. If the gene–environment interaction is replicated in future research, then CRC survivors, particularly those with high-risk genotypes, may benefit from behavioral interventions such as limiting red meat consumption while maintaining a healthful weight to improve their prognosis.
The strengths of this study include its moderately large sample size, the long period of follow-up, and the availability of detailed information on participants’ personal history. Our study also has several limitations. The information on diet and lifestyle habits was self-reported by participants when we started this investigation, thus potentially introducing bias due to misclassification; however, such bias would not have influenced the genetic findings. In addition, colorectal carcinogenesis is a long process during which disease may promote weight loss and patients may modify their food habits. Therefore, retrospective BMI and diet data might not reflect the real-life situation before death in patients with CRC. Future research assessing diet or dietary alterations post-diagnosis is needed to further elucidate possible gene-environment interactions in CRC outcomes.
Conclusions
Overall, our study provides evidence that rs11763517 of the LEP gene, and rs9436301 and rs7602 of the LEPR gene are likely to be associated with CRC survival. The LEP/LEPR-CRC survival association was modified by red meat consumption and BMI. Notably, the SNPs examined in this study were tag SNPs, which are considered to be only indicators for specific regions of interest and thus may not reflect causality47. If our findings are successfully replicated in other well-powered studies, certain variants of the LEP and LEPR genes may serve as novel prognostic biomarkers for CRC, and CRC survivors may improve their prognosis through lifestyle changes.
Supporting Information
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Peizhong Peter Wang, Yun Zhu.
Collected the data: Patrick S. Parfrey, John R. Mclaughlin, Peizhong Peter Wang.
Performed the analysis: Meizhi Du, Yu Wang.
Wrote the paper: Meizhi Du, Yu Wang.
Revised the paper: Meizhi Du, Yu Wang, Jillian Vallis, Matin Shariati, Patrick S. Parfrey, John R. Mclaughlin, Peizhong Peter Wang, Yun Zhu.
Acknowledgements
We express our sincere gratitude to the Canadian Institutes of Health Research Team, the National Cancer Institutes of Health, and Ontario Registry for their contributions to data collection.
Footnotes
↵*These authors contributed equally to this work.
- Received October 13, 2022.
- Accepted March 30, 2023.
- Copyright: © 2023, Cancer Biology & Medicine
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.