Abstract
Long non-coding RNAs (lncRNAs) refer to a group of RNAs that are usually more than 200 nucleotides and are not involved in protein generation. Instead, lncRNAs are involved in different regulatory processes, such as regulation of gene expression. Different lncRNAs exist throughout the genome. LncRNAs are also known for their roles in different human diseases such as cancer. HOTAIR is an lncRNA that plays a role as an oncogenic molecule in different cancer cells, such as breast, gastric, colorectal, and cervical cancer cells. Therefore, HOTAIR expression level is a potential biomarker for diagnostic and therapeutic purposes in several cancers. This RNA takes part in epigenetic regulation of genes and plays an important role in different cellular pathways by interacting with Polycomb Repressive Complex 2 (PRC2). In this review, we describe the molecular function and regulation of HOTAIR and its role in different types of cancers.
keywords
Introduction
The physiological and developmental complications of humans lead us to the point that the limited number of protein-coding genes compared with whole genome cannot explain the complexity of human features1. Most of the genomic DNAs (at least 70%-90%) are transcribed to the RNAs that do not produce any proteins. These parts of the genomes are known as non-coding RNA (ncRNA) genes, which produce efficient RNA molecules1–4.
LncRNAs consist of a group of ncRNAs, including thousands of various species5. These RNAs are usually more than 200 nucleotides and are mostly transcribed by RNA pol II from different regions across the genome. They play different roles such as transcriptional and post-transcriptional regulation inside the cell. Recently, lncRNAs are known as key regulators of gene expression6–8. By working on embryonic stem cells and through analyzing the ribosome profiling data, Chew et al.9 revealed that many lncRNAs are protein-coding contaminants. LncRNAs also regulate the activity of epigenetic machinery during cell differentiation10. In fact, many lncRNAs recruit chromatin-modifying proteins (e.g., PRC2) to specific sites of genome and affect gene expression through regulating chromatin states11.
Based on their roles, the dysregulation of lncRNAs is involved in several diseases including cancer12,13. Du et al.14 analyzed the expression of different lncRNAs in various tumors. Through this analysis, they identified the lncRNAs related to different cancers and their clinical prognosis. Dysregulation of lncRNAs is related to prognosis, metastasis, and recurrence in different cancer types. Studies show that dysregulation of certain lncRNAs affect several processes related to oncogenesis, including cell growth and proliferation15. The over expression of some lncRNAs with proto-oncogenic function in normal cells increases tumor growth and matrix invasion of cancer cells14,16. Moreover, over-expression of oncogenic lncRNAs results in tumor-cell proliferation and metastasis through chromatin looping and some other processes17.
In this review, we describe the oncogenic roles of HOTAIR long-non coding RNA as one of the most important regulatory RNAs in human cells. We also present the molecular function and regulation of this lncRNA in different types of cancer.
HOTAIR lncRNA
HOTAIR lncRNA was introduced by Rinn et al.18 as a spliced and polyadenylated RNA with 2,158 nucleotides and 6 exons. This RNA arises from the transcription of antisense strand of HoxC gene, which is specifically situated between HoxC11 and HoxC12 on chromosome 12q13.13. Computational and Northern blot analysis revealed that HOTAIR does not show any stem loops suggestive of being a pre-miRNA. These analysis also suggested that HOTAIR is preferentially expressed in posterior and distal sites of the human body. In an experiment on 10 mammalian genomes and 3 non-mammalian vertebrates, He et al.19 looked for matches to the 6 exons of HOTAIR and its two conserved domains. They reported a poor sequence conservation and, by contrast, noticeably conserved structures for HOTAIR. They also reported that HOTAIR has evolved faster compared with adjacent HoxC genes.
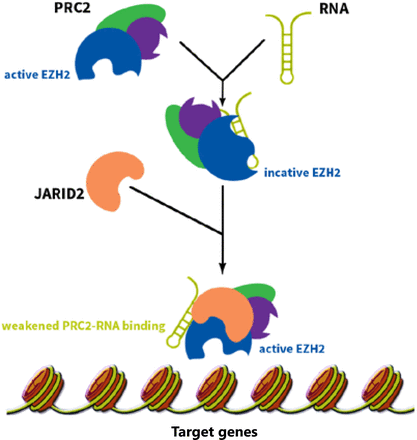
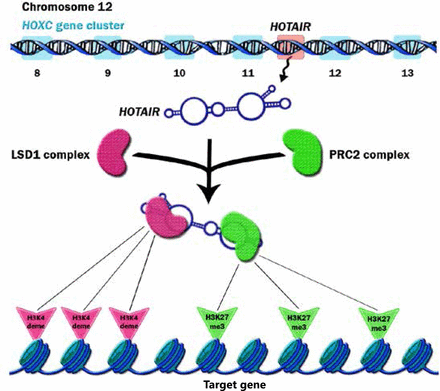
HOTAIR is a trans-acting lncRNA and has different target loci such as HOXD4. HOTAIR interacts with Polycomb Repressive Complex 2 (PRC2) and is necessary for PRC2 occupancy and histone H3 lysine-27 trimethylation of different genes in different chromosomes. PRC2 is a histone methyltransferase that implements epigenetic silencing during different processes including cancer development20. HOTAIR localizes and targets PRC2 genome wide21. PRC2 is a complex that contains three major subunits, including EZH2, SUZ12, and EED. Although EZH2 is the key player for the methyltransfer process, other subunits are also required to regulate EZH2 catalytic activity22. The affinity of EZH2 to RNA is regulated by EED, which increases the specificity of PRC2 function. Cifuentes-Rojas et al.23 investigated the PRC2-RNA interaction precisely. They showed that RNA directs PRC2 to its target gene and simultaneously inhibits the enzymatic activity of EZH2. When PRC2 reaches its target gene, JARID2 binds to EZH2 to impair PRC2’s binding to RNA and thereby activates EZH2’s function (Figure 1). Knockdown of JARID2 results in reduction of H3K27me3 levels on some target genes24. JARID2 also may have a negative impact on PRC2’s function, and deletion of JARID2 results in the enhancement of H3K27me3 levels on some target genes25. HOTAIR functions as a molecular scaffold and interacts not only with PRC2 but also with LSD1 complex to regulate gene expression. LSD1 involves in demethylation of histone H3 at lysine 426,27 (Figure 2). Specifically, PRC2 binds to a 5' domain and LSD1 to a 3' domain of HOTAIR, and HOTAIR coordinates their functions for chromatin modification28. Through these functions, HOTAIR affects the expression of multiple genes involved in various cellular functions21.
The RNAs recruiting PRC2 complex inhibit PRC2 function. These RNAs guide PRC2 to its target gene and inhibits EZH2 enzymatic activity at the same time. When PRC2 reaches its target gene, another protein called JARID2 comes into play and binds to EZH2, weakens EZH2-RNA binding, and consequently activates EZH2’s function.
HOTAIR gene is located on chromosome 12 inside the HoxC locus, specifically between HoxC11 and HoxC12. After the expression of HOTAIR, this lncRNA recruits PRC2 and LSD1 complexes and thus functions as a bridge. HOTAIR directs these complexes to their target genes and as a result regulates the trimethylation of H3K27 and demethylation of H3K4 at targeted genes. H3K27me3 and H3K4deme refer to the trimethylation of histone H3 at lysine-27 and the demethylation of histone H3 at lysine 4, respectively.
HOTAIR lncRNA as an oncogenic factor in different cancers
HOTAIR is an oncogenic factor and can be used as a prognostic biomarker in different cancer types29. HOTAIR lncRNA plays a key role in the initiation and progression of different types of cancer such as cervical cancer and nasopharyngeal carcinoma30,31. HOTAIR also plays an important role in promoting malignancy32. To assess the association between HOTAIR expression levels and lymph node metastasis, Cai et al.33 in a meta-analysis surveyed a total of 748 patients from 8 studies. In this meta-analysis, they showed that the patients with high HOTAIR expression level had a higher incidence compared with that in patients with low HOTAIR expression level. Moreover, Alves et al.34 investigated the role of HOTAIR in epithelial-to-mesenchymal transition (EMT) and also its role in arising and maintenance of cancer stem cells (CSCs). They revealed that HOTAIR plays an important role in the process of tumorigenicity by triggering EMT and acquiring stemness.
In fact, HOTAIR is involved in several processes associated with carcinogenesis such as those affecting the mobility, proliferation, apoptosis, invasion, aggression, and metastasis of the cells (Table 1). PRC2 and LSD1 complexes to exert epigenetic modifications and suppressing a number of genes such as tumor and metastasis suppressor genes. Given these crucial functions, HOTAIR is applied as a potential biomarker of various human cancers. In addition, measuring the expression level of HOTAIR can help us detect the progression stage of cancer and predict the survival possibility of an individual21,27,38,39. Furthermore, HOTAIR is involved in the resistance of cancer cells to cisplatin. This role of HOTAIR is at least attributed to the downregulation of P21 gene. Liu et al.60 found that knockdown of HOTAIR could resensitize the responses of A549/DDP cells to cisplatin. Interestingly, different functional SNPs across whole HOTAIR locus have been reported to influence the cancer risk61.
Overexpression of HOTAIR in different cancers
Regulation of HOTAIR through different pathways
The expression level of HOTAIR gene and the function of its transcript can be controlled by several factors (Table 2). The DNA methylation pattern of downstream intergenic CpG island of HOTAIR may have an important effect on its expression level62. Moreover, the post-synthetic methylation of some cytosines of HOTAIR has been reported. This post-synthetic methylation within or near important functional regions of HOTAIR may play an important role in the regulation of HOTAIR function4.
HOTAIR regulatory factors
The function of HOTAIR can be suppressed by argonaute2 (Ago2) complex in the presence of microRNA-141 (miR-141). MiR-141, unlike HOTAIR, is a suppressor of tumorigenicity, invasiveness, and malignancy in several cancer types. MiR-141 first binds to HOTAIR to suppress it, and Ago2 complex comes into play and cleaves the HOTAIR32.
A type of phosphoglycoprotein called osteopontin (OPN), which is an extracellular matrix protein, can transcriptionally activate and increase HOTAIR expression in cancer cells. Receptor CD44, a positive regulator of OPN, affects the expression level of HOTAIR. By contrast, interferon regulatory factor 1 (IRF1) decreases HOTAIR expression level by binding to its promoter. In fact, OPN regulates IRF1 and affects its signaling pathway, thus activating HOTAIR expression by suppressing the function of IRF163.
The protein c-Myc is another element that impacts the expression of HOTAIR. c-Myc is an oncoprotein that plays a role in the development of several types of cancer through regulating several protein-coding and non-coding genes. c-Myc recognizes a putative E-box element in the upstream region of HOTAIR, which is approximately located at the 1,053 upstream within its promoter. c-Myc directly interacts with this E-box element and upregulates the expression of HOTAIR. In addition, knockdown of c-Myc can reduce both HOTAIR expression and its promoter activity, whereas upregulation of c-Myc gene increases HOTAIR expression and its promoter activity29. Moreover, silico analysis identified four potential Myc-binding sites within HOTAIR promoter4.
Researchers working on human breast cancer cells have shown that diethylstilbestrol and bisphenol-A can upregulate the expression of HOTAIR in these cells66. Some evidence showed the existence of estrogen response elements in the promoter of HOTAIR. Estrogen receptors (ERs) and ER coregulators, such as histone methylases mixed lineage leukemia (MLL) 1, MLL3, and CREB-binding protein/p300, induce the expression of HOTAIR by binding to its promoter64.
TGF-β is another factor that induces HOTAIR expression and involves in EMT, which results in arising and maintenance of CSCs4. Furthermore, evidence suggests the effect of type I collagen (Col-1) on HOTAIR upregulation. Zhuang et al.65 showed that Col-1, which is aberrantly enriched in the tumor microenvironment, can induce the expression of HOTAIR in lung cancer cells53. In addition, HOTAIR overexpression has been reported in non-small cell lung cancer54.
HOTAIR functions
Coordination with PRC2
Li et al.26 showed that directed deletion of HOTAIR lncRNA in mouse can result in activation of hundreds of genes. Different downstream pathways and genes are attributed to the molecular roles of HOTAIR in human cells. HOTAIR can act through promoting the chromatin relocalization done by PRC2. This targeting of PRC2 in the genome leads into a distinct pattern of gene expression necessary for breast cancer progression21,35. Specifically, an overlap exists between HOTAIR-binding motif and BRCA1-binding region in EZH2. This finding indicates that decreased expression of BRCA1 results in elevated recruitment of PRC2 by HOTAIR in breast cancer cell lines4.
HOTAIR functions through Wnt/β-catenin
The key role of HOTAIR in the development and progression of esophageal squamous cell carcinoma (ESCC) has been revealed36. HOTAIR exerts its role through activating Wnt/β-catenin signaling pathway. The Wnt/β-catenin signaling pathway is an important pathway in the development of ESCC. HOTAIR recruits PRC2 directly to the promoter region of Wnt inhibitory factor 1 (WIF-1), leading to the reduction of WIF-1 expression and consequently the activation of Wnt/β-catenin signaling pathway37,61. In addition, different functional SNPs across the whole HOTAIR locus that affects the regulation of HOTAIR may influence ESCC risk61.
Involvement in EMT
Through a series of in vitro and in vivo assays on epithelial ovarian cancer (EOC) tissues, researchers showed that a significant association exists between HOTAIR expression level and metastatic stage of EOC. This association may be due to the regulation of certain matrix metalloproteinases (MMPs) and EMT-related genes by HOTAIR. HOTAIR expression is also associated with FIGO stage/metastasis of lymph nodes; thus, this factor could be a potential biomarker or therapeutic target in EOC patients50.
Inhibition of HOTAIR in gastric cancer cells leads to EMT process reversal and reduction of invasiveness mediated by the expression of MMP1 and MMP367. Some evidence shows that suppression of miR-7 by HOTAIR can mediate EMT progression in breast cancer. This microRNA can inhibit the SETDB1 and STAT3 pathway in breast cells68.
Functions as competitive endogenous RNAs
HOTAIR can function as competitive endogenous RNAs (ceRNAs) in gastric cells by recruiting the microRNAs targeting the HER-2. Thus, HOTAIR and HER-2 may have coexpression in gastric cancer tissues69. Given this recently identified role of HOTAIR, the finding indicative of the positive interaction between HOTAIR and HER2 is worth to research in other types of cancer cells.
miRNA-130a binding sites were found in HOTAIR lncRNA. A negative correlation between HOTAIR and miRNA-130a has been demonstrated in gallbladder cancer tissues compared with nearby normal tissues. Thus, the oncogenic role of HOTAIR is not only by recruiting PRC2 but also partly through negative regulation of miRNA-130a29.
HOTAIR regulates various genes in tumors
HOTAIR overexpression is also related to hepatocellular carcinoma (HCC)40–43. In an experiment, Ding et al.44 showed that suppression of HOTAIR leads to the increase of RNA binding motif 38 (RBM38) proteins, which play a role in the regulation of cell motility. They also showed that RBM38 expression levels were lower in HCC tissues compared with noncancerous tissues in the same patients. Therefore, HOTAIR increases the aggression and invasion of HCC cells by suppressing RBM38 expression.
Some reports indicate that HOTAIR plays an oncogenic role partially via the downregulation of HOXA555. The results also suggest that this lncRNA plays a potential oncogenic role through influencing the expression of specific genes associated with cell adhesion such as MUC5AC and ASTN156. Kogo et al.45 suggested that HOTAIR regulates the expression of multiple genes in cooperation with PRC2 and raises the levels of undifferentiated cancer cells in CRC patients.
In an experiment, Yan et al.48 measured the HOTAIR expression level of Ta/T1 bladder cancer tissues and adjacent normal tissues, which were collected from 110 patients. They reported that 90 specimens had high HOTAIR expression levels, which were inversely correlated with WIF-1 expression. HOTAIR may also be involved in the development of colorectal cancer46,47.
Wu et al.49 reported an increase in HOTAIR expression level in renal carcinoma cells. They also reported that knockdown of HOTAIR by siRNA impacts the cell cycle in the G0/G1 phase and also decreases the cell proliferation and invasion in vitro. These effects resulted in the reduction of HOTAIR binding ability to EZH2 and consequently the reduction of H3K27me3 on HOTAIR target genes. Furthermore, they showed that inhibition of HOTAIR expression resulted in suppression of growth of xenograft tumors formed by renal carcinoma cells. In addition, they demonstrated that inhibition of HOTAIR expression and modulation of covalent histones activated transcriptional state of cell cycle-related gene.
Tang et al.57 investigated the potential roles of HOTAIR in melanoma cells and showed that HOTAIR is overexpressed in metastatic melanoma tissues. They showed that knockdown of HOTAIR by siRNAs resulted in the reduction of motility and invasion of human melanoma cell line A375. They also reported that HOTAIR is involved in promoting gelatinase activity in melanoma cells.
Different studies revealed that increasing the expression of HOTAIR has several effects, such as increasing the proliferation and aggression of cancer cells in pancreatic cancer tissues. On the contrary, downregulation of HOTAIR has opposite effects, including inhibition of cell cycle progression, reducing proliferation, and increasing apoptosis. However, the results of gene array studies showed that some differences exist between HOTAIR-regulated genes in pancreatic cells and breast cancer cells. These studies suggest several interferon- and cell cycle-related genes as targets of HOTAIR in pancreatic cancer cells59. Furthermore, overexpression of HOTAIR may be associated with endometrial carcinoma51,52.
Although the exact roles of HOTAIR in glioma and glioblastoma are still challenging, HOTAIR can be used as a prognostic biomarker in glioma58.
Conclusion
Different studies provide some evidence for the crucial roles of HOTAIR in the initiation and progression of various cancers. Understanding the biological roles of HOTAIR in different types of cancer helps us to determine the efficiency of this lncRNA as a diagnostic or predictive biomarker. However, HOTAIR has been suggested as a biomarker for most cancer types. Thus, conducting a meta-analysis of HOTAIR expression in all cancer types may help in the identification of the cancers that have the highest probability of HOTAIR overexpression. Hence, HOTAIR expression level can be used as a potential prognostic factor for various cancers. In addition, screening the HOTAIR overexpression can help us identify cancer progression and tumor stage. Expression analysis can be conducted by different quantitative techniques such as RT-PCR. These assays would be valuable when the RNA level can be differentially analyzed in samples of patients such as urine, blood, and mucus. However, clinical trials are needed in the future to find this RNA as a suitable biomarker or therapeutic target in cancer.
HOTAIR can also be further considered as a therapeutic target to improve the sensitivity of therapy for different tumors. Moreover, the efficacy of these therapeutic approaches can be further expanded via identification of exact molecular pathways underlying the regulation of HOTAIR expression. Although some regulatory pathways of HOTAIR expression have been reported, thorough identification of those pathways requires more studies and experiments.
Footnotes
Conflict of interest statement No potential conflicts of interest are disclosed.
- Received January 17, 2015.
- Accepted February 12, 2015.
- Copyright: © 2015, Cancer Biology & Medicine
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.