Abstract
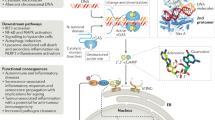
Innate DNA sensing via the cyclic GMP-AMP synthase–stimulator of interferon genes (cGAS–STING) mechanism surveys microbial invasion and cellular damage and thus participates in various human infectious diseases, autoimmune diseases and cancers. However, how DNA sensing rapidly and adaptively shapes cellular physiology is incompletely known. Here we identify the STING–PKR-like endoplasmic reticulum kinase (PERK)–eIF2α pathway, a previously unknown cGAS–STING mechanism, enabling an innate immunity control of cap-dependent messenger RNA translation. Upon cGAMP binding, STING at the ER binds and directly activates the ER-located kinase PERK via their intracellular domains, which precedes TBK1–IRF3 activation and is irrelevant to the unfolded protein response. The activated PERK phosphorylates eIF2α, forming an inflammatory- and survival-preferred translation program. Notably, this STING–PERK–eIF2α pathway is evolutionarily primitive and physiologically critical to cellular senescence and organ fibrosis. Pharmacologically or genetically targeting this non-canonical cGAS–STING pathway attenuated lung and kidney fibrosis. Collectively, the findings identify an alternative innate immune pathway and its critical role in organ fibrosis, report an innate immunity-directed translation program and suggest the therapeutic potential for targeting the STING–PERK pathway in treating fibrotic diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Polysome-seq data have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE165910. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Chen, Q., Sun, L. & Chen, Z. J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Roers, A., Hiller, B. & Hornung, V. Recognition of endogenous nucleic acids by the innate immune system. Immunity 44, 739–754 (2016).
Hu, M. M. & Shu, H. B. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu. Rev. Immunol. 38, 79–98 (2020).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA 106, 8653–8658 (2009).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019).
Sharma, S. et al. Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 (2003).
Fitzgerald, K. A. et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003).
Chan, Y. K. & Gack, M. U. RIG-I-like receptor regulation in virus infection and immunity. Curr. Opin. Virol. 12, 7–14 (2015).
Chen, S. et al. TBK1-mediated DRP1 targeting confers nucleic acid sensing to reprogram mitochondrial dynamics and physiology. Mol. Cell 80, 810–827 (2020).
Gao, D. et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl Acad. Sci. USA 112, E5699–E5705 (2015).
Crampton, S. P. & Bolland, S. Spontaneous activation of RNA-sensing pathways in autoimmune disease. Curr. Opin. Immunol. 25, 712–719 (2013).
Fu, J. et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 7, 283ra252 (2015).
Ng, K. W., Marshall, E. A., Bell, J. C. & Lam, W. L. cGAS–STING and cancer: dichotomous roles in tumor immunity and development. Trends Immunol. 39, 44–54 (2018).
Demaria, O. et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl Acad. Sci. USA 112, 15408–15413 (2015).
Deng, L. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).
Tang, C. H. et al. Agonist-mediated activation of STING induces apoptosis in malignant B cells. Cancer Res. 76, 2137–2152 (2016).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019).
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011).
Xu, P. et al. Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol. Cell 56, 723–737 (2014).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Glück, S. & Guey, B. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Wu, S. et al. HER2 recruits AKT1 to disrupt STING signalling and suppress antiviral defence and antitumour immunity. Nat. Cell Biol. 21, 1027–1040 (2019).
Petrasek, J. et al. STING–IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl Acad. Sci. USA 110, 16544–16549 (2013).
Cerboni, S. et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214, 1769–1785 (2017).
Melki, I. et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140, 543–552 (2017).
Hasan, M. et al. Chronic innate immune activation of TBK1 suppresses mTORC1 activity and dysregulates cellular metabolism. Proc. Natl Acad. Sci. USA 114, 746–751 (2017).
Moretti, J. et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171, 809–823 (2017).
Wu, J., Chen, Y. J., Dobbs, N., Sakai, T. & Liou, J. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216, 867–883 (2019).
Liu, Y. et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014).
Luksch, H. et al. STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144, 254–266 (2019).
Motwani, M. et al. Hierarchy of clinical manifestations in SAVI N153S and V154M mouse models. Proc. Natl Acad. Sci. USA 116, 7941–7950 (2019).
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 (2009).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Donnelly, N., Gorman, A. M., Gupta, S. & Samali, A. The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 (2013).
Dever, T. E. Gene-specific regulation by general translation factors. Cell 108, 545–556 (2002).
Hinnebusch, A. G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 (2005).
Franz, K. M., Neidermyer, W. J., Tan, Y. J., Whelan, S. P. J. & Kagan, J. C. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl Acad. Sci. USA 115, E2058–E2067 (2018).
Mohamed, E. et al. The unfolded protein response mediator PERK governs myeloid cell-driven immunosuppression in tumors through inhibition of STING signaling. Immunity 52, 668–682 (2020).
Sen, T. & Saha, P. Aberrant ER stress induced neuronal-IFNβ elicits white matter injury due to microglial activation and T-cell infiltration after TBI. J. Neurosci. 40, 424–446 (2020).
Lipton, J. O. et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161, 1138–1151 (2015).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Hinnebusch, A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 (2014).
Wek, R. C. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol 10, a032870 (2018).
Inglis, A. J. & Masson, G. R. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl Acad. Sci. USA 116, 4946–4954 (2019).
Harding, H. P., Zhang, Y. & Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 (1999).
Dey, M. et al. Mechanistic link between PKR dimerization, autophosphorylation and eIF2α substrate recognition. Cell 122, 901–913 (2005).
Lee, J. H., Yoon, Y. M. & Lee, S. H. TUDCA-treated mesenchymal stem cells protect against ER stress in the hippocampus of a murine chronic kidney disease model. Int. J. Mol. Sci. 20, 613 (2019).
Li, P. et al. TUDCA attenuates intestinal injury and inhibits endoplasmic reticulum stress-mediated intestinal cell apoptosis in necrotizing enterocolitis. Int. Immunopharmacol. 74, 105665 (2019).
Kusaczuk, M. Tauroursodeoxycholate-bile acid with chaperoning activity: molecular and cellular effects and therapeutic perspectives. Cells 8, 1471 (2019).
Kaur, B. et al. Proteomic profile of 4-PBA treated human neuronal cells during ER stress. Mol. Omics 14, 53–63 (2018).
Zheng, P. et al. 4-PBA improves lithium-induced nephrogenic diabetes insipidus by attenuating ER stress. Am. J. Physiol. Ren. Physiol. 311, F763–F776 (2016).
Zeng, M. et al. 4-PBA inhibits LPS-induced inflammation through regulating ER stress and autophagy in acute lung injury models. Toxicol. Lett. 271, 26–37 (2017).
Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P. & Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332 (2000).
Sanderson, T. H. et al. PKR-like endoplasmic reticulum kinase (PERK) activation following brain ischemia is independent of unfolded nascent proteins. Neuroscience 169, 1307–1314 (2010).
Roberts, Z. J. et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J. Exp. Med. 204, 1559–1569 (2007).
Cerón, S., North, B. J., Taylor, S. A. & Leib, D. A. The STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) stimulates an antiviral state and protects mice against herpes simplex virus-induced neurological disease. Virology 529, 23–28 (2019).
Weiss, J. M. et al. The STING agonist DMXAA triggers a cooperation between T lymphocytes and myeloid cells that leads to tumor regression. Oncoimmunology 6, e1346765 (2017).
Brandizzi, F. & Barlowe, C. Organization of the ER–Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392 (2013).
Shang, G., Zhang, C., Chen, Z. J., Bai, X. C. & Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393 (2019).
Martin, M., Hiroyasu, A., Guzman, R. M., Roberts, S. A. & Goodman, A. G. Analysis of Drosophila STING reveals an evolutionarily conserved antimicrobial function. Cell Rep. 23, 3537–3550 (2018).
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012).
Bennion, B. G. et al. A human gain-of-function STING mutation causes immunodeficiency and gammaherpesvirus-induced pulmonary fibrosis in mice. J. Virol. 93, e01806-18 (2019).
Chevalier, R. L., Forbes, M. S. & Thornhill, B. A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 75, 1145–1152 (2009).
Chung, K. W. et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 30, 784–799 (2019).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008).
Gall, A. et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36, 120–131 (2012).
Ahn, J., Gutman, D., Saijo, S. & Barber, G. N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl Acad. Sci. USA 109, 19386–19391 (2012).
Luo, X. et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology 155, 1971–1984 (2018).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Robichaud, N., Sonenberg, N., Ruggero, D. & Schneider, R. J. Translational control in cancer. Cold Spring Harbor Perspect. Biol 11, a032896 (2019).
Delépine, M. et al. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott–Rallison syndrome. Nat. Genet. 25, 406–409 (2000).
Zhang, P. et al. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22, 3864–3874 (2002).
Sutton, M. A. & Schuman, E. M. Dendritic protein synthesis, synaptic plasticity and memory. Cell 127, 49–58 (2006).
Sharma, V., Ounallah-Saad, H. & Chakraborty, D. Local Inhibition of PERK enhances memory and reverses age-related deterioration of cognitive and neuronal properties. J. Neurosci. 38, 648–658 (2018).
Chang, R. C., Wong, A. K., Ng, H. K. & Hugon, J. Phosphorylation of eukaryotic initiation factor-2α (eIF2α) is associated with neuronal degeneration in Alzheimer’s disease. Neuroreport 13, 2429–2432 (2002).
Eyries, M. et al. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat. Genet. 46, 65–69 (2014).
van ‘t Wout, E. F., Hiemstra, P. S. & Marciniak, S. J. The integrated stress response in lung disease. Am. J. Respir. Cell Mol. Biol. 50, 1005–1009 (2014).
Woo, C. W. et al. Adaptive suppression of the ATF4–CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat. Cell Biol. 11, 1473–1480 (2009).
Meng, F. et al. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol. Cell 81, 4147–4164 (2021).
Vargas, J. N. S. et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell 74, 347–362 (2019).
Xie, X. et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc. Natl Acad. Sci. USA 108, 6474–6479 (2011).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
Korfei, M. et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 178, 838–846 (2008).
Shu, S. et al. Endoplasmic reticulum stress is activated in post-ischemic kidneys to promote chronic kidney disease. EBioMedicine 37, 269–280 (2018).
Meng, X. et al. Endoplasmic reticulum stress promotes epithelial-mesenchymal transition via the PERK signaling pathway in paraquat-induced pulmonary fibrosis. Mol. Med. Rep 24, 525 (2021).
Shi, Y. et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18, 7499–7509 (1998).
Volmer, R., van der Ploeg, K. & Ron, D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl Acad. Sci. USA 110, 4628–4633 (2013).
Girardin, S. E. et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587 (2003).
Eaglesham, J. B., Pan, Y., Kupper, T. S. & Kranzusch, P. J. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS–STING signalling. Nature 566, 259–263 (2019).
Wang, P., Li, J., Tao, J. & Sha, B. The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. J. Biol. Chem. 293, 4110–4121 (2018).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Saitoh, T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA 106, 20842–20846 (2009).
Dobbs, N. et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 (2015).
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5, ra20 (2012).
Zhang, Q. et al. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 19, 362–374 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Acknowledgements
We are grateful to J. Han, J. Shao and Z. Jiang for the reagents and Y. J. Zhang for helpful discussions. This research was sponsored by the NSFC Projects (31830052 and 31725017 to P.X.) and the National Key Research and Development Program of China (2021YFA1301401 to P.X.). Thanks also go to the Life Sciences Institute core facilities of Zhejiang University for technical assistance.
Author information
Authors and Affiliations
Contributions
D.Z. and Y.L. carried out most of the experiments. Y.Z., Q.Z., H.G., S.L., S.C., C.M., C.C., Z.L., Y.X., S.O., X.-H.F., T.L. and L.S. contributed to several experiments and helped with data analyses and discussions. P.X. and D.Z. conceived the study and designed the experiments. P.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Claudio Hetz, Katalin Susztak and Søren Paludan for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 STING inhibits cap-dependent translation through the PERK-eIF2α axis and is independent of the UPR.
(A) Expression of STING but not MAVS inhibited new protein synthesis, as determined by immunoblotting for puromycin in HEK293 cells. The graph shows the relative protein synthesis rate normalized to that of GAPDH. n = 3 independent experiments (mean ± SEM), and P values were indicated by one-way ANOVA with the Bonferroni correction. (B) Global protein synthesis was substantially attenuated by inducing expression of a constitutively active form of STING (R281Q) in DLD1 cells via the Tet-On system, as indicated by the declined levels of puromycin. (C) Translation arrest induced by the ER and oxidative stress was typical in STING-deficient HCT116 cells. (D) Ectopic expression of both NvSTING and DrSTING distinctly induced eIF2α but not IRF3 phosphorylation in HEK293 cells. (E) Phosphorylation of endogenous eIF2α in HEK293 cells was induced by ectopic expression of hSTING, NvSTING, or DmSTING. (F-G) Immunofluorescence imaging revealed that eIF2α was mainly localized in the cytoplasm and partially associated with the ER, which was not aggregated upon STING expression. Scale bars=20 μm. (H) eIF2α phosphorylation, which STING triggered, was declined explicitly in the presence of a PERK inhibitor but not an ER stress inhibitor (TUDCA), TBK1 inhibitor (BX795), or mTOR inhibitor (rapamycin) in HCT116 cells. (I) Expression of STING led to eIF2α phosphorylation in HCT116 cells, which was abolished by PERK inhibitors (iPERK-1 and iPERK-2) but not NF-κB inhibitors (TPCA-1 and JSH-23). (J) Ectopic expression of STING R331A/R334A failed to induce the phosphorylation of both eIF2α and IRF3 in HEK293 cells. (K) Immunofluorescence imaging revealed the partial accumulation of ATF6, an indication of ATF6 activation, by the treatment of UPR activator but nut STING agonist. Scale bars=10 μm. (L) Genetic ablation of STING expression did not affect the activation of UPR signalling, as evidenced by the similar levels of phospho-PERK, phospho-eIF2α, and phospho-IRE1α in WT and STING KO MEFs upon DTT treatment. Data shown in A-L represent three independent experiments.
Extended Data Fig. 2 Activation of STING-PERK-eIF2α signalling precedes that of the STING-TBK1-IRF3 cascade.
(A) In the presence of TBK1 inhibitor, ectopic expression of STING in HEK293 cells failed to induce IRF3 phosphorylation, but its capability to induce eIF2α phosphorylation remained intact. (B) STING activation in primary MEFs, as induced by agonist diABZI, led to PERK-eIF2α signalling activation, as indicated by immunoblotting for phospho-PERK. Data in a and b represent three independent experiments.
Extended Data Fig. 3 STING interacts with and directly activates PERK via its intracellular C-terminal domain.
(A) Coimmunoprecipitation assays showed that the interacting motifs, that is, the C-terminal region of the PERK kinase domain, were sufficient for STING interaction. (B) Immunofluorescence imaging revealed that PERK ICD and its kinase-dead mutant were mainly localized in the cytoplasm and probably associated with cytoskeleton and plasma membrane. Scale bars=10 μm. (C) PERK KO HEK293 cells were generated via a CRISPR-based strategy and verified by immunoblotting for PERK. Data in A-C represent three independent experiments.
Extended Data Fig. 4 The PERK-eIF2α cascade is discrete with TBK1-IRF3 and NF-κB signalling and STING-induced autophagy.
(A) Ectopic expression of STING CTD induced the phosphorylation of endogenous eIF2α in HEK293 cells in a dose-dependent manner. (B) Immunofluorescence imaging revealed that STING CTD was distributed in the cytoplasm and the nucleus, distinct from the full-length STING that associates the ER. Scale bars=20 μm. (C) Treatment of PERK inhibitor unaffected the STING-induced autophagy in DLD1 cells, as indicated by LC3 lipidation. Data in A-C represent three independent experiments. (D-E) PERK inhibition did not substantially regulate STING-induced NF-κB signalling activation, either STING versions from human (D), anemone (E), or Drosophila (E), as evidenced by the reporter assays. In D-E, n = 3 independent experiments (mean ± SEM), and P values were indicated by one-way ANOVA with the Bonferroni correction.
Extended Data Fig. 5 STING-initiated translation program selectively facilitates inflammatory and survival signalling.
(A) The mRNA levels of DDX21 and CDC25B were not notably altered upon the treatment of cGAMP at 1 h, when their protein levels were enhanced. n = 3 independent experiments (mean ± SEM), and P values were indicated by one-way ANOVA with the Bonferroni correction.
Extended Data Fig. 6 The STING-PERK-eIF2α axis is critical for damage-induced cellular senescence.
(A) HU-induced senescence was substantially attenuated upon the treatments of PERK inhibitor but not inhibitors of IKKβ or the UPR, as revealed by SA-β-Gal staining. (B-C) PERK ablation in HCT116 cells via a CRISPR-based strategy significantly suppressed HU-induced senescence, as evidenced by SA-β-Gal staining (A). This suppression was reversed upon reconstitution of murine PERK (B). The KO and reconstitution efficiencies were verified by immunoblotting (bottom panel, B), and statistics were calculated for SA-β-Gal-stained cells (right panel, A). (D) The PERK-deficient cell population was generated by CRISPR-mediated genome editing, and its efficiency was determined by immunoblotting. (E) Perk ablation in primary MEFs led to a decreased expression of p21Waf1, a key indicator of cellular senescence. (F) Reconstitution of STING-deficient HCT116 cells with the 2LA mutant did not restore HU-induced apoptosis, as revealed by endogenous cleaved PARP proteins. Scale bar=100 μm (A, B, C). In A, B, and E, n = 3 independent experiments (mean ± SEM), and P values were indicated by one-way ANOVA with the Bonferroni correction. Data in C, D, and F represent three independent experiments.
Extended Data Fig. 7 Intervening the STING-PERK-eIF2α axis ameliorates the progression of fibrotic pulmonary diseases.
(A) Pulmonary fibrosis was induced by bleomycin in WT and STING KO C57BL/6 mice (top panel). Genetic ablation of STING substantially ameliorated lung fibrosis symptoms, as evidenced by the diminished areas of collagen and declined Ashcroft fibrosis scores. Scale bars=100 μm. Data shown represent mean ± SEM (n = 4 mice in the saline group; n = 8 mice in the other groups), and P values were indicated by one-way ANOVA with the Bonferroni correction. (B-C) Bleomycin-induced pulmonary fibrosis was markedly attenuated in STING-deficient mice (B) or upon the treatment of PERK inhibitor (C), as revealed by immunoblotting for the reduced levels of α-SMA and collagen I. (D) Schematic figure showed the design, generation, and verification of the Eif2ak3 heterozygotes, and the gRNA target sequence (5’-TGGACAGGAGGTGCCTCGTTGGG-3’) was located in the exon 1 of murine Eif2ak3 gene. (E) PERK heterozygotes (Eif2ak3 + /-) were largely normal in appearance and weight and were fertile, while its homozygotes were embryonic lethal. (F) Bleomycin-induced pulmonary fibrosis was markedly attenuated in PERK half-deficiency mice, as revealed by immunoblotting for α-SMA and collagen I. (G) Immunoblottings for α-SMA and collagen I showed that Bleomycin-induced pulmonary fibrosis was severely prevented by PERK inhibition, but not the inhibition of the UPR. Images of lung sections for individual mice in Extended Data Fig. 7 were provided in Supplementary Information. Data in B, C, F and G represent three independent experiments.
Extended Data Fig. 8 The STING-PERK-eIF2α cascade underlies pulmonary and renal fibrosis.
(A-D) Immunofluorescence imaging showed the cGAMP-triggered upregulation of endogenous α-SMA and collagen I proteins in lung sections, which were substantially dampened in PERK half-deficiency mice (A-B), or upon the treatment of the PERK inhibitor iPERK-2 (C-D). Scale bars=100 μm. Data in A-D represent three independent experiments. (E) As expected, the kidney collected from the UUO surgery group on day 14 was dilated. Scale bars=1 cm. Data shown represent n = 13 mice in the sham group; n = 16 mice in other groups. (F) A schematic figure describing the signalling mechanism and function of this non-canonical cGAS-STING-PERK pathway, which occurred at the ER to regulate mRNA translation and preceded to the classical STING-TBK1-IRF3 cascade that regulates mRNA transcription.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables
Supplementary Table 1: List of recombinant DNA. Supplementary Table 2: Antibodies used in the study. Supplementary Table 3: Oligos used in the study. Supplementary Table 4: Quality control of polysome-seq samples
Source data
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western Blots and/or gels.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Zhang, D., Liu, Y., Zhu, Y. et al. A non-canonical cGAS–STING–PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat Cell Biol 24, 766–782 (2022). https://doi.org/10.1038/s41556-022-00894-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00894-z
This article is cited by
-
cGAS-STING, inflammasomes and pyroptosis: an overview of crosstalk mechanism of activation and regulation
Cell Communication and Signaling (2024)
-
Mammalian integrated stress responses in stressed organelles and their functions
Acta Pharmacologica Sinica (2024)
-
Second messenger 2'3'-cyclic GMP-AMP (2'3'-cGAMP): the cell autonomous and non-autonomous roles in cancer progression
Acta Pharmacologica Sinica (2024)
-
Innate immune sensing of lysosomal dysfunction drives multiple lysosomal storage disorders
Nature Cell Biology (2024)
-
Disulfiram ameliorates STING/MITA-dependent inflammation and autoimmunity by targeting RNF115
Cellular & Molecular Immunology (2024)