Abstract
Therapeutic targeting of KRAS-mutant lung adenocarcinoma represents a major goal of clinical oncology. KRAS itself has proved difficult to inhibit, and the effectiveness of agents that target key KRAS effectors has been thwarted by activation of compensatory or parallel pathways that limit their efficacy as single agents. Here we take a systematic approach towards identifying combination targets for trametinib, a MEK inhibitor approved by the US Food and Drug Administration, which acts downstream of KRAS to suppress signalling through the mitogen-activated protein kinase (MAPK) cascade. Informed by a short-hairpin RNA screen, we show that trametinib provokes a compensatory response involving the fibroblast growth factor receptor 1 (FGFR1) that leads to signalling rebound and adaptive drug resistance. As a consequence, genetic or pharmacological inhibition of FGFR1 in combination with trametinib enhances tumour cell death in vitro and in vivo. This compensatory response shows distinct specificities: it is dominated by FGFR1 in KRAS-mutant lung and pancreatic cancer cells, but is not activated or involves other mechanisms in KRAS wild-type lung and KRAS-mutant colon cancer cells. Importantly, KRAS-mutant lung cancer cells and patients’ tumours treated with trametinib show an increase in FRS2 phosphorylation, a biomarker of FGFR activation; this increase is abolished by FGFR1 inhibition and correlates with sensitivity to trametinib and FGFR inhibitor combinations. These results demonstrate that FGFR1 can mediate adaptive resistance to trametinib and validate a combinatorial approach for treating KRAS-mutant lung cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Downward, J. Targeting RAS signalling pathways in cancer therapy. Nature Rev. Cancer 3, 11–22 (2003)
Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013)
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009)
Scholl, C. et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009)
Luo, J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009)
Stephen, A. G., Esposito, D., Bagni, R. K. & McCormick, F. Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014)
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J. & Der, C. J. Drugging the undruggable RAS: mission possible? Nature Rev. Drug Discov. 13, 828–851 (2014)
Lito, P., Rosen, N. & Solit, D. B. Tumor adaptation and resistance to RAF inhibitors. Nature Med. 19, 1401–1409 (2013)
Samatar, A. A. & Poulikakos, P. I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nature Rev. Drug Discov. 13, 928–942 (2014)
Lito, P. et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014)
Sun, C. et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Reports 7, 86–93 (2014)
Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature Biotechnol. 29, 79–83 (2011)
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002)
Morris, E. J. et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013)
Lee, H. J. et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26, 207–221 (2014)
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010)
Villanueva, J. et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18, 683–695 (2010)
Corcoran, R. B. et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2, 227–235 (2012)
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012)
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012)
Kouhara, H. et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89, 693–702 (1997)
Gozgit, J. M. et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol. Cancer Ther. 11, 690–699 (2012)
Guagnano, V. et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 54, 7066–7083 (2011)
Gavine, P. R. et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056 (2012)
Li, F. et al. FGFR-mediated reactivation of MAPK signaling attenuates antitumor effects of imatinib in gastrointestinal stromal tumors. Cancer Discov. 5, 438–451 (2015)
Traer, E. et al. Ponatinib overcomes FGF2-mediated resistance in CML patients without kinase domain mutations. Blood 123, 1516–1524 (2014)
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005)
Shao, D. D. et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014)
Shimizu, T. et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin. Cancer Res. 18, 2316–2325 (2012)
Fellmann, C. et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell 41, 733–746 (2011)
Taylor, J., Schenck, I., Blankenberg, D. & Nekrutenko, A. in Current Protocols in Bioinformatics (eds Baxevanis, A. D. et al. ) Ch. 10, Unit 10.5 (Wiley, 2007)
Chou, T. C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 (2010)
Poirier, J. T. et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 34, 5869–5878 (2015)
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature Protocols 4, 1064–1072 (2009)
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013)
Saborowski, M. et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 28, 85–97 (2014)
Acknowledgements
We thank M. Sánchez-Céspedes, R. Somwar, and H. Varmus for sharing cell lines; S. Tian, J. Ahn, M. Taylor, A. Shroff, and J. Plevy for technical assistance; C. J. Sherr, L. E. Dow, P. Lito, T. Kastenhuber, and J. Leibold for advice on experimental design and/or for editing the manuscript; and other members of the Lowe laboratory for advice and discussions. This work was supported by a program project grant from the National Cancer Institute (S.W.L., N.R.), a grant from the Center of Experimental Therapeutics (S.W.L., N.R.), a Stand Up To Cancer grant from the American Association for Cancer Research (N.R., J.A.E., C.R.), and a Cancer Center Support grant to MSKCC. E.M. was supported by The Jane Coffin Childs Memorial Fund for Medical Research and a K99/R00 grant from the National Institutes of Health/National Cancer Institute. S.W. was supported by the Annette Kade Fellowship from the Watson School of Biological Sciences. R.W. was supported by a Carl-Duisberg Fellowship from the Bayer Foundation. A.L. was supported by an EMBO Long-Term fellowship. E.d.S. received support through the Geoffrey Beene Cancer Research Center. S.W.L. is the Geoffrey Beene Chair of Cancer Biology and a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Contributions
E.M. conceived the project, performed and analysed experiments, and wrote the paper with assistance of all authors. S.W., C.C., and R.W. performed and analysed in vitro experiments. S.W., J.P.M., and E.d.S. performed and analysed in vivo experiments. A.L. helped design and produce the shRNA library. J.T.P. and C.R. provided and analysed patient-derived xenografts. J.F.G., R.B.C., and J.A.E. provided human specimens. N.R. conceived the project, supervised experiments, and wrote the paper. S.W.L. conceived the project, supervised experiments, analysed data, and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
N.R. is a member of the scientific advisory board of Novartis, AstraZeneca, and Chugai Pharmaceutical.
Additional information
Reviewer Information
Nature thanks J. Tyner and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 A synthetic lethal RNA interference screen identifies different MAPK signalling effectors and FGFR1 as sensitizers to MEK inhibition in KRAS-mutant lung cancer cells.
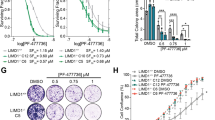
a, Library features and schematic of the TRMPV-Neo vector. b, Schematic outline of the synthetic lethal RNA interference screen for identifying sensitizers to trametinib in KRAS-mutant lung cancer cells. c, Clonogenic assay of KRAS-mutant lung cancer cell lines (H23, H460, and H2030) cultured in the presence of increasing concentrations of trametinib. d, Proliferation assay of H23 and H2030 cells in the presence of increasing concentrations of trametinib for four passages. Data presented as mean of two independent replicates. e, Immunoblot analysis of KRAS-mutant lung cancer cell lines treated with 25 nM of trametinib for 48 h. f, g, Scatter plots illustrating the correlation of normalized reads per shRNA between replicates at the beginning of the experiment (d) and replicates at different time points in the absence (left) or presence (right) of trametinib (25 nM) (e). h, Scatter plot illustrating the fold change in the relative abundance of each shRNA in the library after ten population doublings on doxycycline in the absence or presence of trametinib (25 nM) in H23 cells. Two shRNAs for FGFR1, CRAF, BRAF, and ERK2 were identified as selectively depleted in trametinib-treated cells. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2 Suppression of FGFR1 and different MAPK signalling effectors reduces the proliferation and viability of KRAS-mutant lung cancer cells treated with trametinib.
a, Quantification of fluorescent cells in competitive proliferation assays in H2030 (upper) and A549 (lower) cells transduced with non-targeting control (Ren) or the indicated shRNAs. Data presented as mean (n = 2). Unpaired two-tailed t-test. *P < 0.05, **P < 0.01. b, Immunoblot of H23 and H2030 cells pre-treated with 25 nM trametinib for various times and subsequently treated with 200 nM trametinib for 2 h. c, Immunoblot of H23 cells transduced with doxycycline-inducible shRNAs targeting CRAF and BRAF and treated with trametinib (25 nM) and doxycycline for the times shown. H23 cells were pre-treated with trametinib for 4 days, followed by treatment with doxycycline and trametinib for 4 days. d, Clonogenic assay of H23 cells transduced with BRAF, CRAF, ERK2, and non-targeting control shRNAs, and cultured with DMSO or trametinib (25 nM) for 10 days. Relative growth of DMSO- (grey bars) and trametinib-treated cells (blue and red bars) is shown (right). Data presented as mean ± s.d. (n = 3). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 ERK inhibitor SCH772984 enhances the antiproliferative effects of trametinib in KRAS-mutant lung cancer cells.
a, Clonogenic assay of H2030 (upper) and H460 (lower) cells treated with increasing concentrations of trametinib, ERK inhibitor SCH772984, or their combination as indicated. Percentage inhibition at each concentration of the drugs in H23, H2030, and H460 cells is presented (right). Data presented as mean of three independent experiments (n = 3). b, Immunoblot analysis of H2030 cells treated with trametinib (25 nM), SCH772984 (500 nM), or their combination for the times shown. H2030 cells were pre-treated with trametinib for 4 days, followed by treatment with SCH772984 and trametinib for 2 days. c, Cell viability of H23, H2030, and H460 cells treated with increasing doses of trametinib, ERK inhibitor SCH772984, or their combination for 10 days. Data presented as mean ± s.d. (n = 3). The GI50 was calculated in the absence or the presence of increasing concentrations of SCH772984 (bottom). For gel source data, see Supplementary Fig. 1. Source Data for Extended Data Fig. 3 are available in the online version of the paper.
Extended Data Figure 4 Feedback activation of FGFR1 signalling leads to adaptive resistance to trametinib in KRAS-mutant lung cancer cells.
a, Immunoblot analysis of KRAS-mutant lung cancer cell lines H23 and H2030 treated with 25 nM trametinib for various times. b–d, qRT–PCR for FGFR1 and FGF2 in A549 (b), H2030 (c), and H460 (d) cells treated with trametinib for the indicated times. Data presented as mean normalized for FGFR1 and FGF2 expression ± s.d. (n = 3). e, Immunoblot analysis of A549, H2030, and H358 cells treated with trametinib (25 nM) for various times. f, Quantification of fluorescent cells in competitive proliferation assays in A549, H358, and H460 cells transduced with doxycycline-inducible non-targeting control (Ren) or FGFR1 shRNAs. Data presented as mean ± s.d. (n = 3). g, qRT–PCR for FGFR1 in H23 cells transduced with non-targeting control and FGFR1 shRNAs. Data presented as mean normalized for FGFR1 expression ± s.d. (n = 3). h, Quantification of fluorescent cells in competitive proliferation assays in A549 cells transduced with non-targeting control (Ren) or the indicated shRNAs. Data presented as mean ± s.d. (n = 3). i, qRT–PCR for FGFR2, FGFR3, and FRS2 in A549 cells transduced with non-targeting control, FGFR2, FGFR3, and FRS2 shRNAs. Data presented as mean normalized for FGFR2, FGFR3, and FRS2 expression ± s.d. (n = 3). b–d, Paired two-tailed t-test. f–i, Unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. For gel source data, see Supplementary Fig. 1.
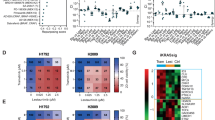
Extended Data Figure 5 Trametinib-induced phosphorylation of FRS2 predicts sensitivity to MEK and FGFR1 combined inhibition.
a, Competitive proliferation assays in the indicated KRAS-mutant cancer cell lines transduced with doxycycline-inducible non-targeting control (Ren) or FGFR1 shRNAs. Data presented as mean ± s.d. (n = 3). b, A panel of lung (H1975, H1650, Ludlu-1, H1703, and H1299), pancreas (MIAPACA, PANC1), and colorectal (SW620, SW480, and DLD1) cancer cell lines were treated with 25 nM trametinib for various times. Lysates were subject to immunoblot analysis with the indicated antibodies. c, Scatter plot illustrating the correlation between depletion of FGFR1 shRNAs-expressing cells and fold change in FRS2 phosphorylation after trametinib treatment in human cancer cells lines (n = 15). d, Representation of the fold change in FRS2 phosphorylation after treatment with trametinib for 12 days in human cancer cell lines (n = 15). a, d, Unpaired two-tailed t-test. c, Two-tailed Pearson’s correlation. **P < 0.01, ***P < 0.001, ****P < 0.0001. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6 Trametinib in combination with ponatinib synergizes at inhibiting cell proliferation of KRAS-mutant lung cancer cells.
a, Serum starved H23 (left) and 3T3 (right) cells were pre-treated with increasing concentration of ponatinib for 24 h (1, 30, 100, and 300 nM), followed by stimulation with FGF2 (50 ng ml−1) for 10 min. Immunoblot analysis for the indicated antibodies is shown. b, Immunoblot analysis of H2030 cells treated with trametinib (25 nM), ponatinib (750 nM), or their combination for the times shown. Cells were pre-treated with trametinib for 4 days, followed by co-treatment with ponatinib and trametinib for 2 days. c, Clonogenic assay of H2030, A549, H2009, and H460 cells treated with increasing concentrations of trametinib, ponatinib, or their combination as indicated. d, Percentage of cell growth inhibition at each concentration of trametinib, ponatinib, or their combination in A549, H2009, and H460 cells after is shown. Data presented as mean of three independent experiments (n = 3). e, Combination index (CI) scores for H23, H2030, A549, H2009, and H460 cells treated with trametinib in combination with ponatinib at the indicated concentrations. Each CI score represents data from at least three independent experiments. For gel source data, see Supplementary Fig. 1. Source Data for Extended Data Fig. 6 in the online version of the paper.
Extended Data Figure 7 Distinct FGFR1 inhibitors sensitize murine and human KRAS-mutant cancer cells to trametinib.
a, Clonogenic assay of a murine lung cancer cell line harbouring KrasG12D and Trp53R270H mutations (KP cell line), and human KRAS-mutant pancreatic cancer cell lines (MIAPACA and PANC1). Tumour cells were cultured with increasing concentrations of trametinib, ponatinib, or their combination as indicated. b, Percentage of cell growth inhibition at each concentration of trametinib, ponatinib, or their combination in KP, MIAPACA, and PANC1 cells is shown. Data presented as mean of three independent replicates (n = 3). c, Combination index scores for KP, MIAPACA, and PANC1 cells treated with trametinib in combination with ponatinib at the indicated concentrations. Each combination index score represents data from at least three independent experiments (n = 3). d, Clonogenic assay of H23, H2030, and H460 cells cultured with increasing concentrations of trametinib alone or in combination with FGFR1 inhibitors BGJ398 (1.5 μM) or AZD4547 (2 μM). e, Percentage of cell growth inhibition at each concentration of trametinib alone or in combination with BGJ398 (1.5 μM) or AZD4547 (2 μM) in H23, H2030, and H460 cells is shown. Data presented as mean of three independent replicates (n = 3). Source Data for Extended Data Fig. 7 is available in the online version of the paper
Extended Data Figure 8 The magnitude of trametinib-induced FRS2 phosphorylation correlates with the sensitivity to trametinib and FGFR1 combined inhibition in human cancer cells.
a, Dot plot illustrating the sensitivity increase to trametinib after the treatment with AZD4547 (2.5 μM) in a panel of KRAS-mutant (n = 15) and KRAS wild-type (n = 15) cancer cell lines. Data presented as mean of two independent replicates (n = 2). b, Scatter plot illustrating the correlation between fold increase in sensitivity to trametinib after treatment with AZD4547 (2.5 μM) or ponatinib (100 nM) and fold change in FRS2 phosphorylation after trametinib treatment in a panel of human cancer cells lines. c, Immunoblot analysis of a panel of human cancer cells treated with trametinib (25 nM) for 6 days. a, Unpaired two-tailed t-test. b, Two-tailed Pearson’s correlation. **P < 0.01, ***P < 0.001, ****P < 0.0001. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 9 Ponatinib prevents trametinib-induced reactivation of MAPK and PI3K signalling. Upregulation of distinct RTKs in KRAS-mutant lung cancer cells after trametinib treatment.
a, Immunoblot analysis of H2030 transduced with PTEN and non-targeting control shRNAs, and treated with trametinib (25 nM) for the times shown. b, Clonogenic assay of H2030 (left) and H460 (middle) cells transduced with PTEN and non-targeting control shRNAs. Cells were treated with ponatinib alone (300 nM) or in combination with trametinib at the indicated concentrations. Quantification of the relative cell growth of H460 cells is shown (right). Data presented as mean of two independent experiments. c, Immunoblot analysis of H2030 transduced with PTEN and non-targeting control shRNAs, and treated with trametinib (25 nM) alone or in combination with ponatinib (750 nM) for the times shown. PTEN suppression did not affect ERK signalling or its inhibition after trametinib treatment but instead activated AKT and, more importantly, attenuated the ability of ponatinib to suppress trametinib-induced increase in pAKT. d, AnnexinV/PI double staining assay of H23 cells treated with vehicle, trametinib (25 nM) alone or in combination with ponatinib (300 nM) or SCH772984 (1 μM) for the times shown (n = 3). e, f, qRT–PCR for EGFR, MET, and ERBB2 in H23 (e) and H2030 (f) cells treated with trametinib for 0, 2, and 4 days. Data presented as mean normalized for EGFR, MET, and ERBB2 expression ± s.d. (n = 3). g, Immunoblot analysis of H23 cells treated with 25 nM of trametinib for various times. h, Immunoblot analysis of serum starved H2030 cells pre-treated with 500 nM or 1 μM of gefitinib, crizotinib, CP-724714, or afatinib for 12 h, followed by stimulation with EGF, HGF, NRG1, or their combination (50 ng ml−1) for 10 min. b, e, f, Unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 10 Unresponsiveness of KRAS-mutant lung cancer cells to MEK inhibitor trametinib is predominantly mediated by feedback activation of FGFR1 signalling.
a, Clonogenic assay of H23 and H2030 cells treated with increasing concentration of trametinib alone or in combination with 500 nM crizotinib, gefitinib, CP-724714, and afatinib, or 300 nM ponatinib. Percentage inhibition at each concentration of the drugs in H23, H460, and H2030 cells is presented (right). Data presented as mean of at least two independent experiments (n = 2). b, CI (combination index) scores for H23, H460, and H23 cells treated with trametinib in combination with crizotinib, gefitinib, CP-724714, afatinib, and ponatinib at the indicated concentrations. Each CI score represents data from at least two independent experiments (n = 2). c, Immunoblot of H23 and H2030 treated with trametinib (25 nM), crizotinib (1 μM), gefitinib (1 μM), CP-724714 (1 μM), and ponatinib (750 nM) for 48 h. d, Immunoblot analysis of H2030 treated with trametinib (25 nM), crizotinib (1 μM), gefitinib (1 μM), CP-724714 (1 μM), ponatinib (750 nM), or their combination for the times shown. Cells were pre-treated with trametinib for 4 days, followed by co-treatment with RTK inhibitors and trametinib for 2 days. For gel source data, see Supplementary Fig. 1. Source Data for Extended Data Fig. 10 are available in the online version of this paper.
Extended Data Figure 11 Suppression of FGFR1 cooperates with trametinib to inhibit growth of KRAS-mutant lung tumours.
a, b, Mice bearing H23 (a) or H2030 (b) xenografts transduced with FGFR1 or non-targeting control shRNAs were treated with either vehicle or trametinib (3 mg/kg body weight). For H23 xenografts, a waterfall representation of the best response for each tumour is shown (n = 8 per group) (a). For H2030 xenografts, the tumour volumes are shown as a function of time after treatment. Error bars, mean ± s.e.m. (n ≥ 4 per group) (b). c, Mice bearing A549 and H2122 xenografts, and JHU-LX55a patient-derived xenograft tumours were treated with vehicle, trametinib (3 mg/kg body weight), ponatinib (30 mg/kg body weight), or both drugs in combination. A waterfall representation of the best response for each tumour is shown (n ≥ 6 per group). d, Body weight of mice bearing A549 xenografts and treated with vehicle, trametinib (3 mg/kg body weight), ponatinib (30 mg/kg body weight), or both drugs in combination for the indicated times (n ≥ 6 per group). e, KrasG12D; Trp53−/− genetically engineered mice harbouring lung adenocarcinomas were treated with vehicle, trametinib (3 mg/kg body weight), ponatinib (30 mg/kg body weight), or both drugs in combination for 7 weeks. A waterfall representation of the response for each tumour after 7 weeks of treatment is shown (n ≥ 5). f, Representative haematoxylin and eosin stains of pancreatic tumour tissue resulting from orthotopic transplantation of GEMM-KPCflox/+ PDAC organoids. Mice were treated with vehicle, trametinib (3 mg/kg body weight), ponatinib (30 mg/kg body weight), or both drugs in combination. Black asterisk indicates necrosis. g, Immunoblot analysis of tumour tissue from mice bearing JHU-LX55a patient-derived xenografts treated with vehicle, trametinib (3 mg/kg body weight), ponatinib (30 mg/kg body weight), or both drugs in combination for 18 days. a–c, e, Unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. For gel source data, see Supplementary Fig. 1. Source Data for Extended Data Fig. 11 are available in the online version of this paper.
Supplementary information
Supplementary Table 1
This table contains primary screening data. Primary data for Figure 1a and Extended Data Figure 1f, g, h. shRNA screen under doxycycline or doxycycline and trametinib 25 nM. (XLSX 633 kb)
Supplementary Table 2
This file contains additional shRNA sequences. shRNA sequences for Figure 2f and Extended Data Figure 4h, i. (XLSX 24 kb)
Supplementary Figure
This file contains full scanned blot images with size markers. (PDF 8302 kb)
Rights and permissions
About this article
Cite this article
Manchado, E., Weissmueller, S., Morris, J. et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651 (2016). https://doi.org/10.1038/nature18600
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18600
This article is cited by
-
Effects of different KRAS mutants and Ki67 expression on diagnosis and prognosis in lung adenocarcinoma
Scientific Reports (2024)
-
Tissue factor overexpression promotes resistance to KRAS-G12C inhibition in non-small cell lung cancer
Oncogene (2024)
-
Establishing a prognostic model based on immune-related genes and identification of BIRC5 as a potential biomarker for lung adenocarcinoma patients
BMC Cancer (2023)
-
Rational combinations of targeted cancer therapies: background, advances and challenges
Nature Reviews Drug Discovery (2023)
-
Scribble mis-localization induces adaptive resistance to KRAS G12C inhibitors through feedback activation of MAPK signaling mediated by YAP-induced MRAS
Nature Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.