Abstract
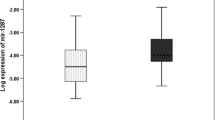
MicroRNAs (miRs) are short non-coding RNAs that bind complementary sequences in mRNA resulting in translation repression and/or mRNA degradation. We investigated expression of the reported metastasis-associated miRs-335, 206, 135a, 146a, 146b, 10b, 21, let7a and let7b in normal mucosa, non-metastatic and metastatic colorectal cancer (CRC). Expression of target miRs in micro-dissected paraffin embedded tissues was evaluated in 15 primary tumours with adjacent normal tissue from patients that were disease-free at 4 years (cohort A) and 19 paired primary tumours with corresponding liver metastases (cohort B) by quantitative real-time PCR. Increased expression of miR-21, mir-135a and miR-335 was associated with clinical progression of CRC, while miR-206 demonstrated an opposite trend. The levels of mir-21 did not associate with the expression of PTEN, an important tumour suppressor in CRC and one of many putative targets of miR-21, but interestingly was associated with stage of disease in the PTEN expressing tumours. Surprisingly, let7a, a KRAS-targeting miR, showed elevated expression in metastatic disease compared to normal mucosa or non-metastatic disease, and only in KRAS mutation positive tumors. Finally, a prognostic signature of miR 21,135a, 335, 206 and let-7a for detecting the presence of metastases had a specificity of 87% and sensitivity of 76% for the presence of metastases. In summary, we have shown stage-associated differential expression of five out of nine tested metastasis-associated miRs. We have further found that an analysis of these five miRs expression levels in primary tumors significantly correlates with the presence of metastatic disease, making this a potential clinically useful prognostic tool.





Similar content being viewed by others
Abbreviations
- miR:
-
MicroRNA
- CRC:
-
Colorectal cancer
- APC:
-
Adenomatous polyposis coli
- FFPE:
-
Formalin fixed paraffin embedded
- DFS:
-
Disease free survival
- LM:
-
Liver metastasis
- LCM:
-
Laser-capture-micro dissection
- RFS:
-
Recurrence free survival
- SNP:
-
Single nucleotide polymorphism
- PTEN:
-
Phosphatase and tensin homolog
References
Parkin DM, Bray F, Ferlay J et al (2005) Global cancer statistics, 2002. CA Cancer J Clin 55(2):74–108
Kurkjian C, Murgo AJ, Kummar S (2008) Treatment of recurrent metastatic colon cancer in the age of modern adjuvant therapy. Clin Colorectal Cancer 7(5):321–324
Eccles SA, Welch DR (2007) Metastasis: recent discoveries and novel treatment strategies. Lancet 369(9574):1742–1757
Iorio MV, Croce CM (2009) MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27(34):5848–5856
Williams AE (2008) Functional aspects of animal microRNAs. Cell Mol Life Sci 65(4):545–562
Kulda V, Pesta M, Topolcan O et al (2010) Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200(2):154–160
Nielsen BS, Jorgensen S, Fog JU et al (2011) High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 28(1):27–38
Schee K, Fodstad O, Flatmark K (2010) MicroRNAs as biomarkers in colorectal cancer. Am J Pathol 177(4):1592–1599
Wang P, Zou F, Zhang X et al (2009) microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 69(20):8157–8165
Schetter AJ, Leung SY, Sohn JJ et al (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299(4):425–436
Nagel R, le Sage C, Diosdado B et al (2008) Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res 68(14):5795–5802
Johnson SM, Grosshans H, Shingara J et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120(5):635–647
Hurst DR, Edmonds MD, Welch DR (2009) Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res 69(19):7495–7498
Asangani IA, Rasheed SA, Nikolova DA et al (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27(15):2128–2136
Tavazoie SF, Alarcon C, Oskarsson T et al (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175):147–152
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449(7163):682–688
Zhang W, Winder T, Ning Y et al (2011) A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol 22(1):104–109
King CE, Wang L, Winograd R et al (2011) LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene 30(40):4185–4193
Rossi L, Bonmassar E, Faraoni I (2007) Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res 56(3):248–253
Peltier HJ, Latham GJ (2008) Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14(5):844–852
Chang GJ, Rodriguez-Bigas MA, Skibber JM et al (2007) Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst 99(6):433–441
Negrini M, Calin GA (2008) Breast cancer metastasis: a microRNA story. Breast Cancer Res 10(2):203
Slaby O, Svoboda M, Fabian P et al (2007) Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72(5–6):397–402
Meng F, Henson R, Lang M et al (2006) Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130(7):2113–2129
De Roock W, Jonker DJ, Di Nicolantonio F et al (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304(16):1812–1820
Chin LJ, Ratner E, Leng S et al (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68(20):8535–8540
Christensen BC, Moyer BJ, Avissar M et al (2009) A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 30(6):1003–1007
Koga Y, Yasunaga M, Takahashi A et al (2010) MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila) 3(11):1435–1442
Song G, Zhang Y, Wang L (2009) MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem 284(46):31921–31927
Adams BD, Cowee DM, White BA (2009) The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol 23(8):1215–1230
Yan D, Dong Xda E, Chen X et al (2009) MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem 284(43):29596–29604
Gabriely G, Wurdinger T, Kesari S et al (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28(17):5369–5380
Zhang JG, Wang JJ, Zhao F et al (2010) MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 411(11–12):846–852
Zhu S, Si ML, Wu H et al (2007) MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 282(19):14328–14336
Vogelstein B, Fearon ER, Hamilton SR et al (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319(9):525–532
Hu X, Schwarz JK, Lewis JS Jr et al (2010) A microRNA expression signature for cervical cancer prognosis. Cancer Res 70(4):1441–1448
Hu Z, Chen X, Zhao Y et al (2010) Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 28(10):1721–1726
Juan D, Alexe G, Antes T et al (2010) Identification of a microRNA panel for clear-cell kidney cancer. Urology 75(4):835–841
Li X, Zhang Y, Ding J et al (2010) Survival prediction of gastric cancer by a seven-microRNA signature. Gut 59(5):579–585
Yanaihara N, Caplen N, Bowman E et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9(3):189–198
Acknowledgments
Research support from the Canadian Institute of Health Research (J.D.), the Ontario Institute for Cancer Research (J.D.) and Ortho-Biotech Canada (J.D.) is greatly appreciated. The authors would like to thank Ms. Colleen Crane for her technical support. Presented in part at the 2010 European Society of Medical Oncology Meeting, Milan, Italy.
Conflict of Interest
All authors declare no conflict of interests with respect to the contents of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
First two authors contributed equally to this study.
Rights and permissions
About this article
Cite this article
Vickers, M.M., Bar, J., Gorn-Hondermann, I. et al. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis 29, 123–132 (2012). https://doi.org/10.1007/s10585-011-9435-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-011-9435-3