Abstract
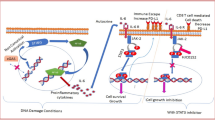
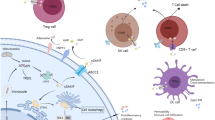
Immune adaptor protein like STING/MITA regulate innate immune response and plays a critical role in inflammation in the tumor microenvironment and regulation of metastasis including breast cancer. Chromosomal instability in highly metastatic cells releases fragmented chromosomal parts in the cytoplasm, hence the activation of STING via an increased level of cyclic dinucleotides (cDNs) synthesized by cGMP-AMP synthase (cGAS). Cyclic dinucleotides 2’ 3’-cGAMP and it's analog can potentially activate STING mediated pathways leading to nuclear translocation of p65 and IRF-3 and transcription of inflammatory genes. The differential modulation of STING pathway via 2’ 3’-cGAMP and its analog and its implication in breast tumorigenesis is still not well explored. In the current study, we demonstrated that c-di-AMP can activate type-1 IFN response in ER negative breast cancer cell lines which correlate with STING expression. c-di-AMP binds to STING and activates downstream IFN pathways in STING positive metastatic MDA-MB-231/MX-1 cells. Prolonged treatment of c-di-AMP induces cell death in STING positive metastatic MDA-MB-231/MX-1 cells mediated by IRF-3. c-di-AMP induces IRF-3 translocation to mitochondria and initiates Caspase-9 mediated cell death and inhibits clonogenicity of triple-negative breast cancer cells. This study suggests that c-di-AMP can activate and modulates STING pathway to induce mitochondrial mediated apoptosis in estrogen-receptor negative breast cancer cells.






Similar content being viewed by others
References
Soysal SD, Tzankov A, Muenst SE (2015) Role of the tumor microenvironment in breast cancer. Pathobiol J Immunopathol Mol Cell Biol 82(3–4):142–152
Sormendi S, Wielockx B (2018) Hypoxia pathway proteins as central mediators of metabolism in the tumor cells and their microenvironment. Front Immunol. https://doi.org/10.3389/fimmu.2018.00040/full
Sachet M, Liang YY, Oehler R (2019) The immune response to secondary necrotic cells. Apoptosis. https://doi.org/10.1007/s10495-017-1413-z
Woo S-R, Corrales L, Gajewski TF (2015) Innate immune recognition of cancer. Annu Rev Immunol 33:445–474
Weinberg SE, Sena LA, Chandel NS (2015) Mitochondria in the regulation of innate and adaptive immunity. Immunity 42(3):406–417
Singh K, Sripada L, Lipatova A, Roy M, Prajapati P, Gohel D et al (2018) NLRX1 resides in mitochondrial RNA granules and regulates mitochondrial RNA processing and bioenergetic adaptation. Biochim Biophys Acta Mol Cell Res 1865:1260–1276
Li A, Yi M, Qin S, Song Y, Chu Q, Wu K (2019) Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J Hematol Oncol. https://doi.org/10.1186/s13045-019-0721-x
Corrales L, McWhirter SM, Dubensky TW, Gajewski TF (2016) The host STING pathway at the interface of cancer and immunity. J Clin Invest 126(7):2404–2411
Basit A, Cho M-G, Kim E-Y, Kwon D, Kang S-J, Lee J-H (2020) The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp Mol Med 52(4):643–657
Medrano RFV, Hunger A, Mendonça SA, Barbuto JAM, Strauss BE (2017) Immunomodulatory and antitumor effects of type I interferons and their application in cancer therapy. Oncotarget 8(41):71249–71284
Lu C, Klement JD, Ibrahim ML, Xiao W, Redd PS, Nayak-Kapoor A et al (2019) Type I interferon suppresses tumor growth through activating the STAT3-granzyme B pathway in tumor-infiltrating cytotoxic T lymphocytes. J Immunother Cancer 7(1):157
Fuertes MB, Woo S-R, Burnett B, Fu Y-X, Gajewski TF (2013) Type I interferon response and innate immune sensing of cancer. Trends Immunol 34(2):67–73
Maimela NR, Liu S, Zhang Y (2018) Fates of CD8+ T cells in tumor microenvironment. Comput Struct Biotechnol J 22(17):1–13
Zhu Y, An X, Zhang X, Qiao Y, Zheng T, Li X (2019) STING: a master regulator in the cancer-immunity cycle. Mol Cancer. https://doi.org/10.1186/s12943-019-1087-y
Andrade WA, Firon A, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA et al (2016) Group B streptococcus degrades cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell Host Microbe 20(1):49–59
Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G (2015) Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol (Baltimore, Md.: 1950) 194(4):1545–1554. https://doi.org/10.4049/jimmunol.1402066
Ablasser A, Hur S (2020) Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol 21:17–29
Archer KA, Durack J, Portnoy DA (2014) STING-dependent type I IFN production inhibits cell-mediated immunity to listeria monocytogenes. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1003861
Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE et al (2013) STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during chlamydia trachomatis infection. mBio. https://doi.org/10.1128/mBio.00018-13
Ahn J, Barber GN (2019) STING signaling and host defense against microbial infection. Exp Mol Med 51(12):1–10
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308
Tomar D, Singh R, Singh AK, Pandya CD, Singh R (2012) TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta—Mol Cell Res 1823(2):316–326
Huber KVM, Olek KM, Müller AC, Soon Heng Tan C, Bennett KL, Colinge J et al (2015) Proteome-wide small molecule and metabolite interaction mapping. Nat Methods 12(11):1055–1057
Bhatelia K, Singh A, Tomar D, Singh K, Sripada L, Chagtoo M et al (2014) Antiviral signaling protein MITA acts as a tumor suppressor in breast cancer by regulating NF-κB induced cell death. Biochim Biophys Acta. https://doi.org/10.1016/j.bbadis.2013.11.006
Chattopadhyay S, Sen GC (2017) RIG-I-like receptor-induced IRF3 mediated pathway of apoptosis (RIPA): a new antiviral pathway. Protein Cell 8(3):165–168
Sun F, Liu Z, Yang Z, Liu S, Guan W (2019) The emerging role of STING-dependent signaling on cell death. Immunol Res 67(2–3):290–296
Vargas-Rondón N, Villegas VE, Rondón-Lagos M (2017) The role of chromosomal instability in cancer and therapeutic responses. Cancers. https://doi.org/10.3390/cancers10010004
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa407
Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, Jacob LS, Patwa R, Shah H, Xu K, Cross JR, Massagué J (2020) Carcinoma–astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. https://doi.org/10.1038/nature18268
Zhang M, Lee AV, Rosen JM (2017) The cellular origin and evolution of breast cancer. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a027128
Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL et al (2018) Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell 71(5):745-760.e5
Su T, Zhang Y, Valerie K, Wang X-Y, Lin S, Zhu G (2019) STING activation in cancer immunotherapy. Theranostics 9(25):7759–7771
Yanai H, Chiba S, Hangai S, Kometani K, Inoue A, Kimura Y et al (2018) Revisiting the role of IRF3 in inflammation and immunity by conditional and specifically targeted gene ablation in mice. Proc Natl Acad Sci 115(20):5253–5258
Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A et al (2017) Signalling strength determines proapoptotic functions of STING. Nat Commun 8(1):1–10
Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, Duran M, Pauli C, Shaw C, Chadalavada K, Rajasekhar VK, Genovese G, Venkatesan S, Birkbak NJ, McGranahan N, Lundquist M, LaPlant Q, Healey JH, Elemento O, Chung CH, Lee NY, Imielenski M, Nanjangud G, Pe’er D, Cleveland DW, Powell SN, Lammerding J, Swanton C, Cantley LC (2018) Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553(7689):467–472. https://doi.org/10.1038/nature25432
Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, Jacob L, Patwa R, Shah H, Xu K, Cross JR, Massagué J (2016) Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533(7604):493–498. https://doi.org/10.1038/nature18268
Kwon J, Bakhoum SF (2020) The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov 10(1):26–39
Acknowledgements
This work was partially supported by the Department of Science and Technology, Govt. of India, Grant Number INT/Korea/P-39 to Prof. Rajesh Singh. Authors acknowledge the instrumentation facility at the Department of Biochemistry, The M. S. University of Baroda, Vadodara. Kritarth Singh received a Senior Research Fellowship from the University Grants Commission (UGC), Govt. of India. We also acknowledge DST FIST for providing an instrumentation facility for the work. Anjali Shinde received her fellowship from ICMR, India. Minal Mane received her fellowship from CSIR, Govt. of India. Dhruv Gohel received his fellowship from ICMR, India. Fatema Currim received her fellowship from INSPIRE, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vasiyani, H., Shinde, A., Roy, M. et al. The analog of cGAMP, c-di-AMP, activates STING mediated cell death pathway in estrogen-receptor negative breast cancer cells. Apoptosis 26, 293–306 (2021). https://doi.org/10.1007/s10495-021-01669-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-021-01669-x