Abstract
Objective: In patients with head and neck squamous cell carcinoma (HNSCC), cetuximab [a monoclonal antibody targeting epidermal growth factor receptor (EGFR)] has been shown to improve overall survival when combined with radiotherapy in the locally advanced setting or with chemotherapy in first-line recurrent and/or metastatic (R/M) setting, respectively. While biomarkers of resistance to cetuximab have been identified in metastatic colorectal cancer, no biomarkers of efficacy have been identified in HNSCC. Here, we aimed to identify biomarkers of cetuximab sensitivity/resistance in HNSCC.
Methods: HNSCC patients treated with cetuximab at the Curie Institute, for whom complete clinicopathological data and formalin-fixed paraffin-embedded (FFPE) tumor tissue collected before cetuximab treatment were available, were included. Immunohistochemistry analyses of PTEN and EGFR were performed to assess protein expression levels. PIK3CA and H/N/KRAS mutations were analyzed using high-resolution melting (HRM) and Sanger sequencing. We evaluated the predictive value of these alterations in terms of progression-free survival (PFS).
Results: Hot spot activating PIK3CA and KRAS/HRAS mutations were associated with poor PFS among HNSCC patients treated with cetuximab in the first-line R/M setting, but not among HNSCC patients treated with cetuximab in combination with radiotherapy. Loss of PTEN protein expression had a negative predictive value among HNSCC patients treated with cetuximab and radiotherapy. High EGFR expression did not predict cetuximab sensitivity in our patient population.
Conclusions: Hot spot activating PIK3CA and RAS mutations predicted cetuximab resistance among HNSCC patients in the first-line R/M setting, whereas loss of PTEN protein expression predicted resistance to cetuximab when combined to radiotherapy.
keywords
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the most common cancer of the head and neck, and the seventh most common cancer overall, affecting around 600,000 patients per year worldwide1. The main risk factors are smoking and alcohol consumption, which are responsible for the majority of HNSCC cases in the oral cavity, pharynx, and larynx. Human papillomavirus (HPV) was also recently identified as a risk factor for oropharyngeal cancer2. Despite advances in multimodal therapy, the overall 5-year survival rate of patients with locally advanced HNSCC remains poor around 40%–50%. Overall survival in the locally advanced setting has been shown to be improved by adding cetuximab to radiation therapy3. In the first-line recurrent and/or metastatic (R/M) setting, cetuximab has been shown to improve overall survival when combined with chemotherapy4. Cetuximab is therefore widely used in both the locally advanced and recurrent and/or metastatic settings5–8. Despite these improvements, only a minority of patients in these settings benefit from cetuximab treatment. Thus, it is critical to identify predictive biomarkers of sensitivity/resistance to cetuximab in HNSCC.
Epidermal growth factor receptor (EGFR) amplifications were not shown to predict cetuximab efficacy in combination with chemotherapy9 and other mechanisms of resistance to EGFR inhibitors have been reported including the activation of downstream signals such as via KRAS and PIK3CA mutations10. A gene signature based on more than 509 differentially expressed genes was reported to be predictive of response to cetuximab in HNSCC11.
In 2015, The Cancer Genome Atlas reported that the molecular landscape of HNSCC includes identified mutations in various oncogenes [PIK3CA (21%) and HRAS (4%)] and tumor suppressor genes [TP53 (72%), CDKN2A (22%), FBXW7 (5%), KMT2D (MLL2) (18%), and PTEN (2%)]12. Phosphoinositide 3-kinases (PI3Ks) play key regulatory roles in multiple cellular processes, including cell survival, proliferation, and differentiation. A broad range of human cancers exhibit frequent alterations in many components of the PI3K/AKT pathway. PIK3CA mutations/amplifications and PTEN loss, respectively, occur in around 34% and 12% of HNSCC cases13.
In metastatic colorectal cancer, KRAS mutations were reported to predict resistance to cetuximab14,15. In cervical cancer patients treated with cetuximab and radiotherapy in a curative intent, downstream PI3K/AKT pathway activation was associated with a resistance to cetuximab16.
In the present study, we aimed to identify the predictive biomarkers of response to cetuximab by analyzing EGFR and PTEN expression, and PIK3CA and RAS mutations.
Patients and methods
Patients and samples
This study included HNSCC patients treated with cetuximab at the Curie Institute, from whom complete clinicopathological data and formalin-fixed paraffin-embedded (FFPE) tumor tissues collected before the cetuximab initiation were available. Disease staging was based on the 7th revised edition (2010) of the American Joint Committee on Cancer (AJCC). All patients were informed that their tumor samples might be used for scientific purposes and had the opportunity to decline. This study was approved by the Internal Review Board of the Curie Institute, and was conducted in accordance with the ethical principles of the Declaration of Helsinki.
DNA extraction
From FFPE tissues, we obtained 6 tissue sections (6-µm thick), and a 7th tissue section that was stained with hematoxylin-eosin. The tumor-rich areas were macrodissected using a single-use blade, and the samples underwent proteinase K digestion in a rotating incubator at 56 °C for 3 days. DNA was extracted using the Nucleospin® 8 Tissue kit (Macherey-Nagel, GmbH & Co. KG, Germany).
RAS and PIK3CA mutations
To screen for mutations, high-resolution melting (HRM) primers were designed for HRAS (exons 2 and 3), NRAS and KRAS (exons 2–4), and PIK3CA (exons 9 and 20). Polymerase chain reaction (PCR) for HRM analysis was performed using the fluorescent DNA-intercalating dye LC green (Idaho Technology), in a 384-well plate using a LightCycler480® (Roche). The reaction mixture had a final volume of 15 µL, and contained LC green, UDP Glycosylase (Roche), and Roche Master Mix (Roche). The reaction conditions were as follows: 40 °C for 10 min, 95 °C for 10 min; 50 cycles of 95 °C for 15 s, 55–65 °C for 15 s, and 72 °C for 25 s; followed by 95 °C for 1 min, and then melting from 65 °C to 95 °C, rising 0.02 °C per s. All samples were tested in duplicate. HRM analysis was performed using Genescan software (Roche). All samples, including the wild-type exons, were plotted on a differential plot graph according to their melting profiles. When an abnormal HRM curve was suspected, the samples were sequenced using the Sanger sequencing approach.
HPV genotyping
HPV status was assessed at the Pathology Department, where HPV typing was conducted using total DNA isolated from FFPE samples of HNSCC tumors. Real-time PCR was performed with Sybr® Green and specific primers for HPV16, 18, and 33, using a 7900HT Fast Real-Time PCR System (Applied Biosystems). HPV L1 amplicons from HPV16-, 18-, and 33-negative samples were sequenced by the Sanger method using the GP6+ primer. HPV type identification was performed via sequence alignment with HPV reference sequences using the NCBI nucleotide blast program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Immunohistochemistry
Immunohistochemical assays using EGFR antibodies (monoclonal mouse, 31G7, Trypsin; InVitrogen) and PTEN antibodies (monoclonal, PTEN amino acids 321–336; Zymed® Laboratories) were performed at the Pathology Department. From the FFPE samples of HNSCC tumors, 3-µm sections were cut, and were deparaffinized and rehydrated through a series of xylene and ethanol washes. All immunostaining processes were performed using a LEICA (BOND III) automated immunostaining device. IHC was performed in some samples exhibiting no intensity (0) and high intensity (3) for EGFR and PTEN, and a score (0%–100%) for the expression of these proteins was established. Membrane EGFR staining was taken into account. For PTEN, an internal control was a positive expression of PTEN in the stromal fibroblasts. A normal prostate tissue was used for an external control in every manipulation. Interpretation of the staining determined the percentage of tumoral positive cells, the intensity of the staining on a three-tiered scale (form 1 to 3) as well as the good quality of the external control and the presence of the internal control. Nuclear expression of PTEN was not considered in the framework of the interpretation of PTEN expression levels. Samples were rated as having high or low expression of EGFR and PTEN using a semi-quantitative histological score: low EGFR = intensity ≤ 2 or protein expression ≤ 80%; high EGFR = intensity > 2 and protein expression > 80%; low PTEN = intensity ≤ 1 and protein expression ≤ 20%; high PTEN = intensity >1 and protein expression > 20%.
Statistical analysis
We recorded patients’ characteristics and survival data up to June 2016. Categorical variables were compared using a Chi-square (χ2) test, with Yates’s correction when appropriate. Patients alive at the end of follow-up were censored at the date of their last visit. Progression-free survival (PFS) was defined as the period from the first day of cetuximab therapy to the date of first disease progression or death from any cause. Survival distributions were estimated by the Kaplan–Meier method, and compared using the log-rank test. The univariate Cox proportional hazard model was used to evaluate the predictive values of clinical and molecular markers for PFS. Results are presented as hazard ratio (HR) and 95% confidence interval (CI). A P value of < 0.05 was considered to be significant. Analyses were performed using R software version 3.3.2 (R Core Team 2016).
Results
Patient and tumor characteristics
We analyzed tumor samples from 118 HNSCC patients who were treated with cetuximab between 2006 and 2015. Three patients were lost to follow-up and thus excluded from the analyses. Table 1 summarizes the clinical, biological, and pathological characteristics of the 115 remaining HNSCC patients. Median follow-up was 54.1 months (range: 3.2–92.5 months). The median age at diagnosis was 60 years. Patients were mainly males. Most patients were smokers. Fifteen patients (13%) had HPV-positive tumors. Of the 115 patients, 77 (67%) were treated with cetuximab in the first-line R/M setting (group 1), and 38 (33%) were treated with cetuximab and radiotherapy in the locally advanced setting (group 2) (Supplementary Tables S1 and S2). Only patients who were not eligible for high-dose cisplatin were treated with cetuximab in combination with radiotherapy in the locally advanced setting. These two groups showed similar distributions of clinical, biological, and pathological parameters, except for age and HPV status (Supplementary Table S3). Compared to group 1, patients from group 2 were older and more frequently had HPV-positive tumors.
Clinical, biological, and pathological characteristics of the 115 HNSCC patients, and their associations with PFS
Predictive value of PIK3CA and RAS mutations on cetuximab efficacy
Whole population
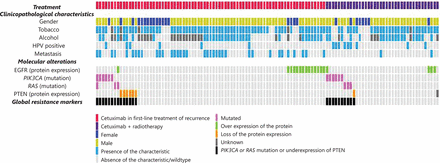
Of the 115 patients, 17 (14.8%) had PIK3CA and/or RAS gene mutations, including 12 (10.4%) with a PIK3CA mutation, 2 (1.7%) with a KRAS mutation, and 4 (3.5%) with a HRAS mutation. One patient had both a PIK3CA mutation and a HRAS mutation (Table 2, Figure 1). All tumors exhibited a wild-type NRAS gene. Supplementary Table S4 presents details of the mutational profiles. In the whole population, PIK3CA, KRAS, and HRAS mutations did not correlate with PFS (Table 2).
Prevalence of DNA mutations and variations in protein expression in the 115 HNSCC samples in relation with PFS
Clinical, biological, pathological, and molecular characteristics of 115 HNSCC patients.
Group 1: cetuximab given in the first-line recurrence setting
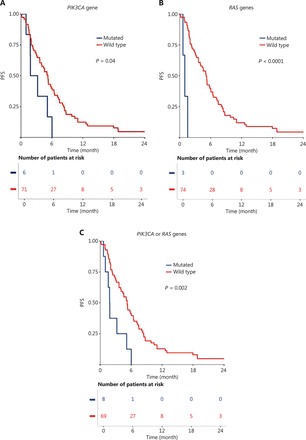
Among the 77 HNSCC patients treated with cetuximab in the first-line R/M setting, the median follow-up was 42.7 months (range: 3.2–57.1 months). Within this group, 7.8% had PIK3CA mutations, and 3.9% had mutations in RAS genes (KRAS or HRAS) (Table 2). PIK3CA mutation (P = 0.04), RAS gene mutation (P < 0.001), and all mutations together (PIK3CA or KRAS or HRAS; P = 0.002) correlated with a poor PFS (Table 2; Figure 2A, 2B, and 2C).
Progression-free survival (PFS) with cetuximab in first-line recurrent and/or metastatic setting, compared between patients with wild-type and oncogene mutations for the following genes: (A) PIK3CA gene, (B) RAS gene, and (C) PIK3CA or RAS genes.
Group 2: cetuximab combined with radiotherapy
Among the 38 HNSCC patients treated with cetuximab combined with radiotherapy, the median follow-up was 62.7 months (range: 4.6–92.5 months). In this group, 15.8% of patients showed hot spot activating PIK3CA mutations, and 7.9% showed RAS gene mutations (Table 2). PFS did not significantly differ between patients with and without mutations of the tested oncogenes.
Predictive value of the loss of PTEN protein expression for cetuximab efficacy
PTEN protein expression loss was detected in 7 patients (6.1%) in the whole population—including 6 patients (7.8%) from group 1, and 1 patient (2.6%) from group 2 (Figure 1, Table 2). Loss of PTEN protein expression was associated with poor PFS in the whole population, although this association was not statistically significant (P = 0.10). The only patient who was treated with cetuximab and radiotherapy and who had a loss of PTEN expression experienced an early recurrence. We observed mutual exclusion between hot spot activating PIK3CA or RAS mutations and loss of PTEN protein expression (Figure 1).
Predictive value of combined oncogene mutations and loss of PTEN protein expression for cetuximab efficacy
Survival analyses suggested a negative predictive value of PIK3CA or RAS mutational status and for loss of PTEN protein expression for cetuximab efficacy (Table 2). At least one resistance biomarker was detected in 24 patients (20.9%) from the whole population—including 14 (18.2%) from group 1, and 10 (26.3%) from group 2 (Table 2).
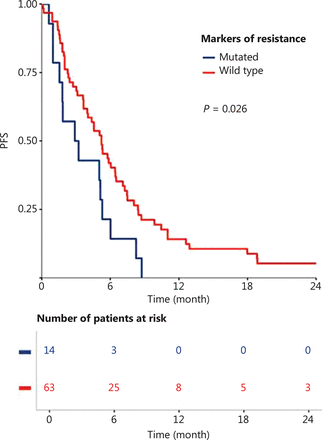
Global resistance biomarker (PIK3CA/RAS mutations and PTEN loss) status significantly correlated with PFS among patients treated with cetuximab in the first-line R/M setting (P = 0.026; Figure 3). These global resistance biomarkers were not significantly correlated with PFS in the subgroup of patients treated with cetuximab alone (Supplementary Figure S1A) or cetuximab in combination with chemotherapy (Supplementary Figure S1B), however, a tendency towards poor prognosis was observed in the cetuximab-treated patients. In this patient subpopulation (group 1), global resistance biomarker status was not significantly associated with any clinical, biological, or pathological parameters, except for age (Supplementary Table S5). Global resistance biomarker status was also found not to be associated with PFS in the whole population, or in the subgroup of patients treated with cetuximab in combination with radiotherapy (group 2; Supplementary Figure S2).
Progression-free-survival (PFS) with cetuximab in first-line recurrence and/or metastatic setting, compared between patients with and without a biomarker of resistance (PIK3CA mutation, RAS mutation, or loss of PTEN protein expression).
Global resistance biomarker status tended to correlate with PFS within the HPV-negative subgroup of patients (Supplementary Figure S3A) as compared to HPV-positive patients (Supplementary Figure S3B).
Predictive value of EGFR protein expression for cetuximab efficacy
EGFR protein overexpression was observed in 18 patients (15.7%) in the whole population, including 14 patients in group 1 (18.2%), and 4 patients (10.5%) in group 2 (Figure 1, Table 2). PFS did not differ significantly according to the EGFR protein level in the whole population, or in the two subgroups (Table 2).
Discussion
HNSCC patients do not undergo any molecular selection prior to treatment with cetuximab, in contrast with metastatic colorectal cancer patients who are required to have a tumor with wild-type KRAS13. EGFR protein expression was not shown to be predictive of cetuximab efficacy in our patient population. The literature reports that EGFR expression and amplifications are not predictive of EGFR inhibition in HNSCC17. The main reason for this is that RAS mutations are rare in HNSCC. The PI3K/AKT pathway is frequently activated in HNSCC, and may constitute a source of tumor escape during EGFR targeting18,19. Therefore, here we focused on alterations in the PI3K/AKT and RAS/MAPK pathways as potential biomarkers of resistance to cetuximab among the 115 HNSCC patients treated with cetuximab at the Curie Institute.
Our patient population was representative of previously reported HNSCC populations, in which tobacco and HPV are main prognostic factors2. Hot spot activating PIK3CA and H/KRAS mutations were observed in 10.4% and 3.5% of HNSCC patients, respectively, which is also consistent with previous reports12,13. IHC revealed loss of PTEN protein expression in 6.1% of our patient population, as compared to a rate of 12% found by RNA sequencing in TCGA12.
We demonstrated that hot spot activating PIK3CA and KRAS/HRAS mutations were associated with poor PFS among HNSCC patients treated with cetuximab in the first-line recurrent setting, but not among patients treated with cetuximab in combination with radiotherapy. We also showed that the loss of PTEN protein expression tended to have a negative prognostic impact in HNSCC patients. EGFR expression did not appear to be a predictive biomarker of cetuximab efficacy, in accordance with previous reports9.
In non-small cell lung cancer (NSCLC), EGFR tyrosine kinase inhibitors show limited efficacy over time, with all patients eventually progressing despite treatment with these drugs20. Activating KRAS mutations and PTEN loss reportedly lead to PI3K/AKT pathway activation, independent of EGFR tyrosine kinase status, driving the downstream cancer survival pathways and supporting resistance21. In patients with HER2-positive breast cancer, PIK3CA mutations reportedly confer resistance to trastuzumab, a monoclonal antibody targeting HER222. Additionally, PTEN loss is frequently reported in HER2-overexpressing esophageal cancer, and has been associated with poor efficacy of trastuzumab-based therapy23. However, studies have reported contradictory data regarding the predictive values of PTEN and PI3K pathway alterations with regards to trastuzumab efficacy24–26.
The findings of our group and others suggest that combined therapy is important for overcoming treatment resistance due to PI3K/AKT pathway alterations in HNSCC. Preclinical studies have investigated the combination of EGFR inhibitors and PI3K/AKT pathway inhibitors, showing synergistic effects that induce apoptosis27. In phase I clinical trials, the pan-class I PI3K inhibitors pilaralisib (SAR245408, XL147) and SAR245409 (XL765) were safely combined with erlotinib, but exhibited limited antitumor activity28,29. A randomized phase II trial demonstrated that combination of the irreversible PI3K inhibitor PX-866 with cetuximab did not improve outcomes among HNSCC patients treated without molecular preselection30. Two ongoing trials are evaluating the combination of BKM120 with erlotinib (NCT01487265) and gefitinib (NCT01570296). Another ongoing phase Ib/II trial is testing the efficacy of the PI3K inhibitor copanlisib in combination with cetuximab among patients with recurrent and/or metastatic HNSCCs that harbor a PIK3CA mutation/amplification and/or a PTEN loss (NCT02822482). It remains to be demonstrated whether these combinations will successfully overcome primary resistance to EGFR inhibitors alone.
Our present results indicated that hot spot activating PIK3CA and RAS mutations were biomarkers of cetuximab resistance among HNSCC patients in the first-line recurrence setting, as is also the case in colorectal cancer14,15. We also showed that PTEN loss might be a biomarker of resistance to cetuximab when administered in combination with radiotherapy. Of note, as all patients treated with either cetuximab or in combination with chemotherapy were treated in the same clinical setting (i.e. R/M), we decided to pull all these patients together in order to get more statistical power. This might have biased the results as prolonged survival on the combination might either be due to cetuximab alone or to chemotherapy or radiotherapy. These results need to be further validated in independent cohorts taking into account treatment regimens. Overall, our results suggest that alterations in the PIK3CA, RAS, and PTEN genes may serve as biomarkers for patient selection in future clinical trials involving treatment with cetuximab alone or in combination with other therapies.
Supporting Information
Acknowledgments
This work was supported by the Fondation ARC pour la recherche sur le cancer (ARC), the Comité d’évaluation et suivi des projets de recherche de transfert of Institut Curie (CEST), and ICGEx project ANR-10-EQPX-03 (Equipement de biologie intégrative du cancer pour une médecine personnalisée). ARC and CEST contributed to the funding for reagents and manpower for the data generation, collection, and interpretation. ICGEx contributed to funding for the sequencing and data interpretation.
Footnotes
Conflicts of interest statement Professor Le Tourneau has participated on the advisory boards of MSD, BMS, Merck Serono, Astra Zeneca, Novartis, Roche, and Nanobiotix.
- Received May 15, 2019.
- Accepted August 28, 2019.
- Copyright: © 2020, Cancer Biology & Medicine
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.