Abstract
A postmenopausal patient with a diagnosis of estrogen receptor (ER) (+), progesterone receptor (PR) (+), and human epidermal growth factor receptor-2 (HER2) (-) breast cancer was reported. The patient refused surgery and was resistant to conventional chemotherapy regimens. Computed tomography and the circulating tumor cell test indicated that the patient’s tumor burden increased rapidly even after several chemotherapy sessions. Multiple genetic aberrances in the phosphatidylinositol3-kinases (PI3K) signaling pathway were detected via next-generation sequencing (NGS)-based liquid biopsy, including a p. G1007R missense mutation in exon 21 of PIK3CA (33.61%), a p.L70fs frameshift mutation in exon 3 of phosphatase and tension homolog deleted on chromosome ten (PTEN) (49.14%), and a p. D1542Y missense mutation in exon 32 of mammalian target of rapamycin (mTOR) (1.66%). Therefore, only the mTOR inhibitor everolimus was administered to the patient. Partial remission (PR) was observed after 2 months, and sustained stable disease (SD) was observed after a year and a half. Subsequent sequencing showed that the mutation ratio of PIK3CA decreased to 4.17%, and that the PTEN and mTOR mutations disappeared, which revealed the significant curative effect of everolimus. We report the first case of successful monotherapy treatment using everolimus in a patient with advanced breast cancer bearing mutations in genes involved in the PI3K/ARK/mTOR signaling pathway. The success of this case highlights the invaluable clinical contribution of NGS-based liquid biopsy, as it successfully provided an optimal therapeutic target for the patient with advanced breast cancer.
keywords
Introduction
Breast cancer is the most frequently diagnosed carcinoma among women. According to data collected by the International Agency for Research on Cancer (IARC) in 2012, 1.7 million women were newly diagnosed with breast cancer, and 521, 900 patients died of the disease, which accounted for 15% of all of the cancer deaths in women1,2. In China, breast cancer is the most common cancer among women, with an estimated 272, 700 new cases and 61, 600 deaths in 20123.
In clinical practice, the treatment of metastatic breast cancer is mainly palliative and relies on chemotherapy, endocrine therapy, and trastuzumab-based targeted therapy4,5. Genetic variations in the human epidermal growth factor receptor 2 (HER2) and other hormone receptors are the basis for the therapeutic treatments for breast cancer patients and the foundation of personalized medicine6. Herceptin (trastuzumab), a recombinant humanized monoclonal antibody (mAb), is the first commercialized drug that targets HER2 on the surface of breast cancer cells7; this is either used as a first-line of treatment, in addition to paclitaxel, or as a second-line monotherapy. Herceptin has shown considerable clinical benefit in patients with early-stage and metastatic breast cancer8,9. However, approximately 15% of patients with breast cancer had HER2 gene amplification or overexpression of the HER2 protein10. For 85% of the HER2 (-) breast cancer patients, information regarding verified biomarkers, which is currently limited, could be used to direct the utilization of targeted medicine.
Everolimus, which is an oral mammalian target of rapamycin (mTOR) inhibitor, has an antitumor effect on different cancers, including breast cancer and renal cell carcinoma, based on multiple clinical trials11,12. After the successful BOLERO-2 study, the use of everolimus and exemestane in patients with estrogen receptor (ER) (+) metastatic breast cancer was approved by the FDA2,13. In clinical investigations and other ongoing clinical trials, everolimus is often used in combination with hormonal drugs, such as exemestane, letrozole, or tamoxifen, to counter endocrine therapy resistance2,14,15. A retrospective study showed that everolimus is more effective in ER (+), PR (+), and HER2 (-) patients with bone and visceral metastases than endocrine monotherapy. The remission rate of the patients treated with everolimus was >80%, whereas that of the patients that received endocrine monotherapy was 60%–70% 16. However, the use of everolimus should be limited owing to its severe adverse effects, such as stomatitis, rash, non-infectious pneumonitis, diarrhea, and hot flushes17. Therefore, we need to determine which breast cancer patients should be treated with everolimus to obtain a higher clinical efficiency with less adverse effects.
Recently, several driver genes involved in the PI3K/AKT/mTOR signaling pathway have been a topic of interest for scientists and clinicians, which involves cell proliferation and apoptosis and tumor pathogenesis and metastasis18. Activation of the phosphatidylinositol3-kinases (PI3K)/AKT/mTOR signaling pathway is common in patients with breast cancer. PIK3CA mutations are detected in 20%–40% of breast cancer patients19, AKT1 mutations are detected in 3% of breast cancer patients, and PTEN mutations occur in approximately 5% of breast cancer patients. Additionally, the activation of mutations in PIK3CA occurs most frequently in patients with ER (+) and HER2 (-) breast cancer (45%)20. The activation of the PI3K/AKT/mTOR signaling pathway is frequently associated with resistance to conventional anticancer therapies, including chemotherapy, radiotherapy, endocrine therapy, and tyrosine kinase inhibitor (TKI)-based targeted therapy21,22, all of which are routine therapeutic treatments for hormone receptor (HR) (+) and HER2 (-) patients. Therefore, it is necessary to determine whether patients bear mutations in genes associated with the PI3K/AKT/mTOR signaling pathway, as this information is essential for the identification of appropriate therapeutic regimens.
An amplification refractory mutation specific-PCR (ARMS-PCR) assay is a routine technique that can detect known mutations in target genes; this technique involves 2 primers, one complementary to the normal allele and the other complementary to a variant at the 3′ end23. The standard ARMS-PCR assay has been frequently utilized to detect hotspot mutations in cancer driver genes, such as epidermal growth factor receptor (EGFR) and KRAS in lung cancer24. However, ARMS-PCR is low-throughput and has low sensitivity for unknown variants, which limit its application for multiple-gene testing. Next-generation sequencing (NGS) is a new paradigm for the massively multiplexed sequence analysis of DNA and RNA. The procedure generates hundreds of millions of short DNA reads (100–250 nucleotides), and all of the reads are sequenced in parallel, which simultaneously provides sequence information for more than 10 million nucleic acids in a sample25,26. NGS approaches not only detect base substitutions but also simultaneously identify insertions and deletions and copy number alterations and translocations. NGS-based comprehensive profiling of all coding regions of the genes of interest is effective for the screening of tumor driver gene mutations in both tissues and blood samples27. Additionally, this approach can detect mutations not detected by the standard ARMS-PCR assay in cancer patients28. Therefore, this study aimed to investigate the clinical significance of NGS in screening possible ER (+), PR (+), and HER2 (-) patients who bear multiple mutations in genes involved in the PI3K/AKT/mTOR signaling pathway and are more responsive to everolimus therapy.
The present study presents the case of an advanced HER2 (-) breast cancer patient who was resistant to conventional chemotherapy regimens. Multiplex genetic mutations involved in the PI3K/AKT/mTOR signaling pathway, including a p. G1007R missense mutation in exon 21 of PIK3CA (33.61%), a L70 frameshift mutation in exon 3 of phosphatase and tension homolog deleted on chromosome ten (PTEN)(49.14%), and a p.D1542Y missense mutation in exon 32 of mTOR (1.66%), were detected via NGS from a plasma circulating tumor DNA (ctDNA) sample, which implied the systemic hyperactivation of the PI3K/AKT/mTOR pathway and potentially high sensitivity to everolimus treatment. Everolimus-based monotherapy was prescribed, and the patient experienced a rapid partial response (PR) to the treatment within 2 months; sustained stable disease (SD) was observed after a year and a half. Additionally, repeated NGS-based liquid biopsy revealed that the mutation ratio of PIK3CA decreased to 4.17%, and that the PTEN and mTOR mutations disappeared. In HER2 (-) breast cancer patients who are resistant to treatment, multiplex genetic mutations in the PI3K/AKT/mTOR pathway can be used as genetic biomarkers to predict positive clinical responses to everolimus, and NGS-based liquid biopsy is an effective genetic assay that can screen potentially actionable mutations in patients with HER2 (-) refractory breast cancer.
Case report
In October 2015, a 52-year-old woman was admitted in Tianjin Medical University Cancer Institute and Hospital. The patient presented with a lump (5 cm × 5 cm) in the upper outer quadrant of the right breast and an extensive ulcer on the right axilla and upper arm, accompanying by reddish discharges, skin infiltration, and edema on the right side of the trunk, just above the navel. Severe swelling occurred in the right upper limb that caused progressive difficulty with movement. The patient did not take any medications, and she did not have any family history of breast cancer. She was first diagnosed with breast cancer in June 2013, when an immobile mass (2 cm × 2 cm) in the right breast grew rapidly in last 2 years before diagnosis, and a significant concentration of radionuclides in situ was confirmed via positron emission tomography-computed tomography (PET-CT). However, no therapeutic regimens was prescribed, because the patient refused any further examination or treatment.
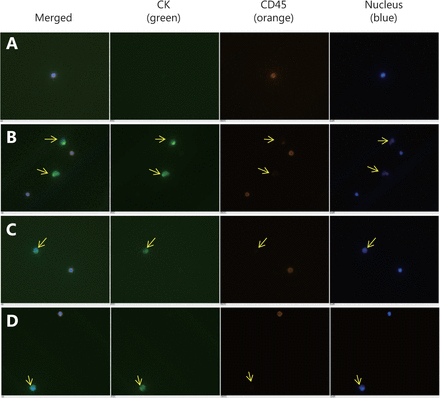
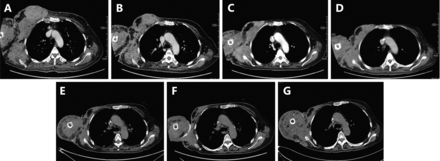
After admission, the biopsy samples were assessed via hematoxylin–eosin staining, immunohistochemistry (IHC), and a fluorescence in situ hybridization (FISH) assay by Breast Pathology Department of Tianjin Medical University Cancer Institute and Hospital. The final diagnosis was invasive carcinoma, and the following molecular profiling features were observed: ER (+): 90%; PR (+): 60%; HER2: (±); Ki-67: 30%; CK5/6: 2%; and EGFR: <1%. HER2 amplification was not identified via FISH. The serum concentration levels of the tumor markers, including CA15-3 (>300 U/mL), CEA (73.5 μg/L), and CA125 (366.9 U/mL), were extremely high. A high amount of circulating tumor cells (CTCs) were also detected, with an average of 31 tumor cells per 2 mL of peripheral blood, which suggested extensive metastasis, in addition to the severe primary tumors ( Figure 1). Enhanced computed tomography (CT) imaging revealed that the cancerous tissues encompassed an area of 8.9 cm × 5.3 cm in size, along with multiple lymphadenectasis in the right axilla, mediastinum, and supraclavicular and infraclavicular regions (Figure 2A). A liver metastatic nodule (2.6 cm × 2.0 cm) was also observed. However, bone metastasis was not detected via emission CT.
Microscopic images of circulating tumor cells (original magnification ×100). Anti-EpCAM was used to capture the cells. Arrows refer to the circulating tumor cells in 2 mL of peripheral blood that tested positive for CK (green) and DAPI (blue) and negative for CD45 (orange). (A) Circulating tumor cells were not detected in the control patient. (B–D) A total of 31 circulating tumor cells were observed in the patient, which suggested a heavier tumor burden.
Images of primary and metastatic lesions on the aortic arch level during the period of disease surveillance. (A) Enhanced computed tomography upon admission showed massive cancerous nodes, and the largest one was 8.9 cm × 5.3 cm in size, which is accompanied by multiple lymphadenectasis and severe edema. (B) After 2 cycles of ET regimen chemotherapy, the patient was stable based on the evaluation via CT (7.1 cm × 4.0 cm) on December 2015. (C) After 2 months of treatment with everolimus, a remarkable remission of the primary lesion (2.8 cm × 2.2 cm) and metastatic lymph node was observed via CT on April 2016, and the clinical response reached PR. (D-G) CT images obtained on August 2016, December 2016, April 2017, and August 2017 showed that the tumor size decreased, and the patient was in stable condition. All therapeutic effect evaluations were carried out according to the RECIST criteria.
As the patient refused biopsy of either the primary breast lump or the liver metastatic nodule, a liquid biopsy was performed to harvest a circling tumor DNA (ctDNA) sample for the NGS assay. The average coverage in the NGS assay was 10000×, and 163 target genes were detected using a commercially available capture-based sequencing panel, LungPlasma panel (Burning Rock Biotech Ltd, Guangzhou, China). The mutation ratios of different genes were calculated as the allelic fractions of the mutations and the frequency of sequencing reads bearing the non-reference allele. Multiplex genetic mutations were detected along the PI3K/AKT signaling pathway, including a p. G1007R missense mutation in exon 21 of PIK3CA (33.61%), a L70 frameshift mutation in exon 3 of PTEN (49.14%), and a p.D1542Y missense mutation in exon 32 of mTOR (33.61%). These mutations, including the missense mutation of PIK3CA within the kinase domain and the loss-of-function mutation of PTEN, a well-known tumor suppressor, are known as hyperactivators of the PI3K/AKT/mTOR pathway. Somatic mutations involving other driver and cancer suppressor genes were also observed, including a p.Q1106H missense mutation in exon 28 of EGFR (15.22%), a p.P1443S missense mutation in exon 17 of WNT signaling pathway regulator (APC) (18.58%), and a p.P151T missense mutation in exon 5 of tumor protein p53 (TP53) (52.72%). These results suggested a systemic hyperactivation of the PI3K/AKT/mTOR signaling pathway, rather than the ErbB receptor tyrosine kinase/ mitogen-activated protein kinase/ extracellular regulated protein kinases (ERBB/MAPK/ERK) pathway in cancer patients.
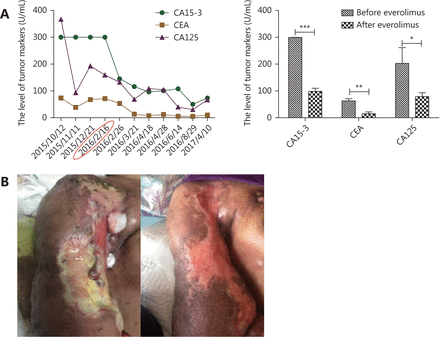
During patient admission, 2 cycles of epirubicin and epirubicin (ET) (day 1: 110 mg of epirubicin and 240 mg of paclitaxel) combination chemotherapy regimens were carried out to reduce the tumor burden and alleviate the rapidly progressing swelling. In addition, a stable tumor (7.1 cm × 4.0 cm) was identified, according to the RECIST criteria, and evaluated via CT scan on December 23, 2015 (Figure 2B). However, the patient experienced grade III myelosuppression and could not tolerate the side effects. Then, the therapy was converted to the vinorelbine and carboplatin (NP) [days 1 and 8: 40 mg of vinorelbine; day 1: area under the plasma concentration-time curve (AUC) = 700 mg of carboplatin] regimen. Unfortunately, the disease progressed rapidly within 2 months following chemotherapy, and the patient refused conventional hormone therapy and preferred the everolimus-based targeted therapy. Upon obtaining informed consent from the patient, only everolimus (10 mg/d) was administered to the patient, beginning February 16, 2016. A macroscopic recession of the primary lesion was observed after 10 days. The clinical response was evaluated via CT scan on April 18, 2016, which was 2 months after the treatment, and a remarkable remission of the primary lesion (2.8 cm × 2.2 cm) was observed (Figure 2C). The clinical response reached PR. The serum concentration level of the tumor markers significantly decreased (Figure 3A). Additionally, her general condition improved significantly, as reflected by the Karnofsky Performance Scale (KPS) score, which increased from 60 to 90. The skin infiltration on the right arm and axilla was in remission. The ulcers healed, and the size of the metastatic nodules on the axilla decreased significantly (Figure 3B). The adverse effects of everolimus, such as fatigue, anorexia, and digestive tract problems, were observed during the therapy, all of which were tolerable.
Changes in the indicators during the disease course. (A) The level of tumor markers was extremely high upon admission, decreased rapidly, and remained at a low level after everolimus treatment since February 16, 2016. CA15-3, CEA and CA125 decreased significantly after the treatment of everolimus (P<0.001,P<0.01,P<0.05). The detectable upper limit of CA15-3 is 300 U/mL, which means more than or equal to 300 U/mL on the graph. (B) The left part presents the condition of the patient before the treatment, with axillary metastatic nodules accompanied by extensive skin infiltration and ulcer. After everolimus treatment, the metastatic nodules disappeared, and the ulcer has healed as shown in the right picture. *P<0.05, **P<0.01, ***P<0.001
The patient was in a stable condition until August 2017, based on regular checkups and CT scans (Figures 2D–G), and the level of the serum tumor markers remained low (Figure 3A). On August 22, 2017, the second NGS-based liquid biopsy was performed. Results showed that the PTEN and mTOR mutations disappeared, and the mutation ratio of the p.H1047R missense mutation in exon 21 of PIK3CA was 4.17%, showing that everolimus significantly inhibited the growth of the cancer cells with mutations in the PI3K/AKT/mTOR signaling pathway.
Discussion
To the best of our knowledge, the current study presents the first report of a breast cancer patient with mutations in genes involved in the PI3K/AKT/mTOR signaling pathway who was successfully treated with targeted therapy alone. Breast cancer cells bearing somatic mutations along the PI3K/AKT/mTOR signaling pathway had strong sensitivity to everolimus29. The BELLE-2 trial compared the use of the pan-PI3K inhibitor buparlisib with fulvestrant or placebo in patients with ER (+) and HER2 (-) metastatic breast cancer, who were resistant to treatment with an aromatase inhibitor. A better progression-free survival was observed in the patients treated with the combination of fulvestrant and buparlisib. This improvement was particularly significant in the patients with mutant PIK3CA detected in their ctDNA30. A phase 1 study of a single-agent AKT inhibitor, ipatasertib, in 52 pretreated patients with various tumor types, including breast cancers, showed that many patients with disease stabilization had PI3K/AKT pathway-activating alterations in their tumors31. Furthermore, the LOTUS trial investigated the addition of ipatasertib to paclitaxel as a first-line therapy for triple-negative breast cancer (TNBC); the combination of ipatasertib and paclitaxel as a first-line therapy for TNBC in the patients bearing PIK3CA/AKT1/PTEN-mutated tumors showed an encouraging progression-free survival of 0.44 and an increase of 4.1 months in the median progression-free survival (median: 9.0 months in the ipatasertib group vs. 4.9 months in the placebo group)32. All of the data suggested that the clinical benefit of PI3K/AKT/mTOR inhibitors may be limited to tumors with mutational activation of PI3K/AKT/mTOR pathway. Moreover, in this case, the condition of the patient improved after chemotherapy. However, durable disease control and systematic benefit were observed after everolimus administration. The rapidity of this response was most likely due to the effect of everolimus, which precisely and fortuitously targeted the genomic alterations in the tumor. Both the clinical trial and case report showed that patients bearing alternations along the PI3K/AKT/mTOR pathway had significant responses to the relevant targeted therapy, and the activation of the PI3K/AKT/mTOR pathway may be a strong predictor of the target therapy.
NGS-based basket and umbrella trials have presented new treatment options for patients who refuse standard therapies. Basket trials refer to trials that aim to evaluate whether a biomarker is predictive of a response to a targeted drug, regardless of the tumor histology33,34. Except in breast cancer cases, therapies that target a mutation along the PI3K/AKT/mTOR pathway had positive effects in other cancers. Two hepatocellular carcinoma patients had genomic alterations that putatively activated the PI3K/AKT/mTOR pathway, as presented by NGS, and received therapies including an mTOR inhibitor; the tumor size of both patients decreased by 30% and 15%, respectively35. A spindle cell neoplasm patient bearing a KIAA1549-BRAF gene fusion, a homozygous deletion of PTEN, and frameshift insertion/deletions in CDKN2A A68fs*51, SUFU E283fs*3, and MAP3K1 N325fs*3, as shown via NGS, had a 25% reduction in the tumor size after combination therapy, which consisted of sorafenib, temsirolimus, and bevacizumab36. In all of the cases, patients with different types of cancer but the same genetic aberrances may respond to the same targeted therapy, as supported by basket trials.
Traditionally, clinical diagnosis and treatment are performed based on the tumor tissues obtained from single biopsy or surgical operation, which causes an underestimation of the intratumor heterogeneity (ITH), leading to an unsatisfactory anti-cancer therapy. A study reported that 63%–69% of all somatic mutations are not detected across different tumor regions37. Additionally, only 5 of 32 somatic non-synonymous coding mutations in the metastasis sites of lobular breast cancer were detected in the primary tumor of the same patient, indicating inconsistency between the primary and metastatic lesions38. Compared to tissue-derived DNA, circulating tumor DNA (ctDNA) has comprehensive genetic features and avoids the intrinsic bias of a solid biopsy, because ctDNA contains DNA from all possible sources, such as orthotropic, circulating, and metastatic tumor cells39.
However, the analysis of ctDNA is challenging because of its characteristics, including a high degree of fragmentation, a low concentration in circulation, and high concentrations of background wild-type DNA40. Previous studies have verified that NGS can detect rare variants with high fidelity, throughput, and speed, which allowed us to understand the characteristics of several cancer genomes and to identify genetic alterations that are common in a variety of human tumor types. Based on this result, targeted capture-based DNA sequencing can focus on a customized region of the genome, which is clinically pragmatic and allows for deeper sequencing coverage41. As previously mentioned, the disadvantages of ctDNA are its sequencing techniques, and ctDNA can be a surrogate for the entire tumor genome.
The NGS-based ctDNA scheme has been a topic of interest for researchers. Recently, a phase 1 clinical trial on the feasibility of NGS-based ctDNA genetic testing, known as liquid biopsy, was conducted with a large cohort of metastatic cancers. Results showed that the specificity of NGS-based liquid biopsy was above 95%, and the sensitivity was 55% in patients with at least one mutation42. In another case, whole-exome sequencing of breast cancer tissue with a PTEN p.H259fs*7 frameshift mutation was performed. In addition, the same variant with a low allele frequency that was initially identified in the tumor tissue was also detected via targeted NGS-based liquid biopsy analysis, which suggested that a higher depth of sequencing coverage is required for the detection of genomic alterations in a liquid biopsy43. Furthermore, the application of liquid biopsy in the detection of epidermal growth factor receptor (EGFR) driver mutations in the blood samples of lung cancer patients has been proven to be an effective method for the detection of multiple genetic mutations in patients without tissue biopsy44. In the present study, we used a capture-based NGS targeted sequencing method to screen all of the exons of 163 genes that are highly mutative and involved in cancer development and progression, and multiplex genetic mutations along the PI3K/AKT/mTOR signaling pathway were observed, which are associated with cancer growth and metastasis. Everolimus was prescribed based on this information, and significant curative effects were observed. Data showed that NGS-based ctDNA genetic testing can be effective in profiling cancer genomic variants and monitoring their changes.
In conclusion, we proposed that multiple-gene testing, particularly that focused on the PI3K/AKT/mTOR pathway, should be performed in order to confirm the choice of everolimus-related adjuvant therapy. Besides, NGS-based liquid biopsy is feasible for the screening of optimal target drugs for ER (+) and HER2 (-) breast cancer or TNBC, especially for refractory advanced breast cancer patients to conquer chemotherapy resistance.
Footnotes
↵* These authors have contributed equally to this work.
Conflict of interest statement No potential conflicts of interest are disclosed.
- Received December 24, 2017.
- Accepted June 1, 2018.
- Copyright: © 2018, Cancer Biology & Medicine
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.