Abstract
Abstract: Epithelial ovarian cancer (EOC) is the leading cause of death among all gynecological malignancies. Despite the technological and medical advances over the past four decades, such as the development of several biological markers (mRNA and proteins biomarkers), the mortality rate of ovarian cancer remains a challenge because of its late diagnosis, which is specifically attributed to low specificities and sensitivities. Under this compulsive scenario, recent advances in expression biology have shifted in identifying and developing specific and sensitive biomarkers, such as microRNAs (miRNAs) for cancer diagnosis and prognosis. MiRNAs are a novel class of small non-coding RNAs that deregulate gene expression at the posttranscriptional level, either by translational repression or by mRNA degradation. These mechanisms may be involved in a complex cascade of cellular events associated with the pathophysiology of many types of cancer. MiRNAs are easily detectable in tissue and blood samples of cancer patients. Therefore, miRNAs hold good promise as potential biomarkers in ovarian cancer. In this review, we attempted to provide a comprehensive profile of key miRNAs involved in ovarian carcinoma to establish miRNAs as more reliable non-invasive clinical biomarkers for early detection of ovarian cancer compared with protein and DNA biomarkers.
keywords
Introduction
Epithelial ovarian cancer (EOC) is the leading cause of death in women among all gynecological malignancies, accounting for about 5% of all cancers and 4.2% of all cancer deaths in women worldwide1,2. Recent prevalence data suggested that it is the most common type of ovarian cancer. EOC is characterized as a heterogeneous group of disease mainly divided into four different histologic subtypes, such as serous, mucinous, clear cell, and endometrioid carcinoma3,4. These complexities are further enhanced by the fact that each subtype has distinct molecular and genetic profiles and behaves and responds differently to treatment, thereby imposing a great hurdle for designing a common therapeutic regimen to treat this devastating illness.
Despite the advances in detection and cytotoxic therapies, only a modest increase in the survival rate beyond five years after initial diagnosis of ovarian cancer has been achieved. The high mortality of patients suffering from this disease is attributed to various factors, including the lack of any specific symptoms in early stages of ovarian cancer; late diagnosis, which poses difficulty in designing an intervention; and the development of chemoresistance in cancer cells. Therefore, improved screening strategies for EOC detection at early stage, as well as effective treatment for advanced stages of ovarian cancer patients, are necessary. In addition, novel biomarkers, particularly a distinct and specific biomarker for each subtype of ovarian cancer that can help in detection at the earliest stage, should be searched and developed, which will serve for designing better and more concentrated treatment strategy for ovarian cancer. Unfortunately, despite comprehensive and multifaceted scientific efforts worldwide, development of biomarkers with ideal specificities and absolute sensitivities remains a challenge. During the last few decades, several attempts to develop serum-based biomarkers for early detection of ovarian cancer have gained limited success because of their low sensitivities and specificities. Therefore, identifying and developing an alternative, non-invasive, sensitive, and more specific biomarker to detect ovarian cancer at the initial stages is necessary for the improved management of ovarian cancer patients. Fortunately, despite the limitations, recently discovered small RNAs, termed microRNAs (miRNAs), have the potential to serve as ideal non-invasive biomarkers for ovarian cancer.
Current status of clinical applications of some important biomarkers for ovarian cancer
The poor diagnosis of ovarian cancer is mainly attributed to the lack of symptoms in the early stages of the disease, which can lead to the occurrence of distant metastasis. Incessant technological advancements in genomics and proteomics have identified multiple molecular biomarkers for ovarian cancer diagnosis. These biomarkers are broadly categorized into gene-based, protein-based, and miRNA-based biomarkers. Gene-based biomarkers measure inherited mutations, epigenetic changes (primarily DNA methylation and histone modifications), and gene expression levels of multiple genes involved in gene regulation. Hereditary germline mutations in genes, such as breast cancer 1/2 (BRCA1 and BRCA2) and DNA mismatch repair genes, primarily hMLH1 and hMSH2, have been detected in at least 10% of all EOCs. Therefore, genetic testing for mutations of these genes may help in potentially identifying patients with higher risk of developing ovarian cancer and designing intervention strategies to reduce the risk of ovarian cancer. Given the development of microarray technology, gene expression profiling enables the rapid comparison of gene expression between normal and malignant cells and identification of genes that are differentially regulated during cancer development.
Carbohydrate antigen 125 (CA-125) is the most widely used protein-based clinical biomarker for the diagnosis of ovarian cancer. Elevated CA-125 serum levels have been detected in about 80%-85% of women with advanced ovarian cancer, but only 50% of patients with stage I ovarian cancer will have an elevated CA-125 level, suggesting that it is neither sufficiently sensitive nor specific for early detection for ovarian cancer. Therefore, CA-125 is mostly used to follow up the stage-wise progression of ovarian cancer in patients with established ovarian cancer5; other important protein biomarkers for ovarian cancers include leptin, prolactin, osteopontin, and insulin-like growth factor II (IGF-II)6.
Another important protein biomarker for early detection of ovarian cancer is human epididymis protein 4 (HE4), a secreted glycoprotein product of the WFDC2 gene. Elevated levels of HE4 have been reported in over 50% of ovarian cancer patients whose tumors do not express CA-125, indicating that HE4 has better potential in monitoring ovarian cancer compared with CA-125. HE4 also has better capacity to distinguish benign and malignant tumors, so it is more useful in identifying early stages of diseases than CA-125, which is also not cancer-specific and can even be elevated in various benign conditions. Many ovarian cancer screening research studies have established that combinatorial biomarker strategy (i.e., multiplexed biomarker approach implying a subset of markers) is more reliable, sensitive, and specific than using a single protein biomarker7. For example, a recent study identified the 5-marker panel for early detection of ovarian cancer that includes five serum biomarkers, namely, macrophage-stimulating protein alpha, tissue inhibitor of metalloproteinases-4, platelet-derived growth factor receptor alpha (PDGF-R alpha), osteoprotegerin, and CA-125. This panel showed an AUC of 0.98 compared with that of CA-125 alone (0.87) and correctly identified 100% ovarian cancer and 95% healthy control samples, providing the total correct agreement of 96.6%8.
A particular disadvantage associated with protein biomarkers is their limited diagnostic value because of their low sensitivity and specificity, high cost, and inconvenience. Therefore, novel and more specific biomarkers with greater diagnostic value should be identified and developed, particularly for the early detection of EOC to minimize the mortality rate and improve survival in ovarian cancer patients. Fortunately, recent evidence indicated that miRNA biomarkers can cope with the flaws of protein biomarkers and serve as more reliable biomarkers for EOC.
MiRNAs: a novel and more reliable biomarker for early detection of ovarian cancer
MiRNAs are a novel class of small (18-24 nucleotides long), evolutionarily conserved non-coding RNA molecules that control various physiological and pathological functions by regulating gene expression at the post-transcriptional level9,10. miRNAs were originally discovered in Caenorhabditis elegans, and they are found in genomes of most eukaryotes, including humans11-13. Recent studies have suggested that miRNAs account for about 1%-5% of the human genome and regulate at least 30% of protein-coding genes14-18. To date, about 940 distinct miRNA molecules have been identified within the human genome19-22.
To improve the survival rate in ovarian cancer patients, highly specific and more sensitive biomarkers should be discovered and developed for ovarian cancer screening and early detection, as well as for survival prediction by assisting monitoring of response to chemotherapeutic drugs. For this purpose, miRNAs as biomarkers offer great tools to evaluate cancer development. They play essential roles in nearly all cellular pathways governing human malignancies, such as carcinogenesis, cancer progression, cell invasion and metastasis, cell survival, and response to therapeutic drugs. In addition, miRNAs also act as oncogenes and tumor-suppressor genes; thus, they have a large potential to serve as promising biomarkers for EOC23,24. Furthermore, miRNA biomarkers are more sensitive and specific than any other biomarkers examined for the diagnosis and prognosis of EOC.
Genomic units of miRNAs and their roles in ovarian cancer: special focus on miR-200 and let-7 families
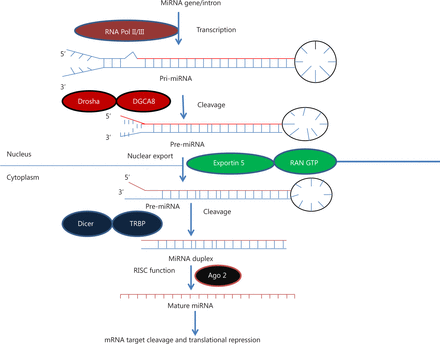
All miRNA genes have well-defined transcriptional units in the genomes. In mammals, miRNAs may be found at multiple locations, such as within introns and exons of protein coding genes, intergenic regions, noncoding genes, repetitive regions, and transposable elements, and they are transcribed similar to mRNAs25. miRNAs located in intergenic regions or in other annotated genes are in antisense orientation, whereas intragenic miRNAs can be oriented in both sense and antisense directions26. Interestingly, about 50% of annotated human miRNAs are associated with cancer and located at fragile sites, which are areas of the genome associated with cancer (Figure 1)27,28.
MicroRNA biogenesis. MiRNA genes are transcribed by RNA polymerase II or III to generate pre-miRNA and cleavage of the pre-miRNA by the microprocessor complex Drosha-DGCR8 (Pasha) in the nucleus to form precursor miRNA. Precursor miRNA is exported from the nucleus to cytoplasm by Exportin-5-Ran-GTP. In the cytoplasm, the RNase Dicer, in complex with the double-stranded RNA-binding protein TRBP, cleaves the pre-miRNA hairpin to its mature length. The miRNA-RISC complex then associates with target mRNA, resulting in repression of the target gene by promoting mRNA degradation and/or translational repression.
Given that this article does not cover the genomic units of all miRNAs and their alterations in ovarian cancer, most of our discussion will largely be confined to the miR-200 and let-7 family of miRs. Thus, only the genomic units of these two miRs are described. The miR-200 family of miRNAs, which are abundantly expressed in epithelial tissues, comprises five members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429). These miRNAs are arranged in two clusters along the human genome. MiR-200a, miR-200b, and miR-429 are clustered on chromosome 1, whereas miR-200c and miR-141 are located on chromosome 1229. Several studies have reported changes in the expression of various members of the miR-200 family and suggested their possible roles in the pathogenesis of ovarian cancer. The miR-200 family is among the most significantly overexpressed miRNAs in EOC. Up-regulated expression of miR-200a and miR-200c has been reported in three types of ovarian cancers: serous, endometrioid, and clear cell carcinomas. However, miR-200b and miR-141 are up-regulated only in endometrioid and serous subtypes, suggesting that the role of the miR-200 family in ovarian carcinoma is more complicated and diverse than initially thought30.
The let-7 (lethal-7) family of miRNA, consisting of 13 different miRNAs, is located and distributed on nine different chromosomes in humans31,32. The expression of the let-7 family is significantly reduced in multiple human cancers. Low expression level of let-7 is associated with poor survival of cancer patients33,34. The let-7 family of miRNAs suppresses multiple ovarian cancer oncogenes, such as KRAS, HRAS, c-MYC35, and HMGA-2 36. Notably, the genomic region harboring let-7a-3/let-7b locus was deleted in 44% of ovarian cancer samples studied. However, restored expression of let-7b significantly reduced ovarian tumor growth both in vitro and in vivo.
MiRNAs are critical regulators of ovarian cancer from pathogenesis to cancer cell survival
MiRNAs perform diverse functions by modulating a broad range of gene expression patterns during development and tissue homeostasis, as well as in the pathogenesis of disease conditions. In ovarian cancer, miRNAs are involved in various cellular functions ranging from carcinogenesis, cell cycle, apoptosis, proliferation, invasion, and metastasis to development of chemoresistance. The role of miRNAs in ovarian cancer has been explored mainly through expression profiling of miRNAs in different cancer types. Various recent studies have reported global and individual miRNA expression patterns in different types of cancers. The first study linking miRNAs and cancer was reported in chronic myelogenous leukemia, in which miR-15 and miR-16 are deleted or down-regulated in tumor patients37. After this pioneer study, various research groups across the globe started to evaluate the role of miRNAs and their deregulations in various cancers. The deregulation of cancer-related miRNAs is due to several factors, including chromosomal rearrangements, alterations in genomic copy numbers, epigenetic modifications, abnormal maturation pathways and their regulation by transcription factors, and miRNA-miRNA interactions38. miRNA deregulation has been reported in almost all type of cancers, including ovarian cancer. Various miRNA species are differentially expressed among different histotypes in ovarian cancer39. These aberrantly expressed miRNAs promote tumor development by inactivating tumor suppressor genes and/or activating oncogenes, a common feature of all ovarian cancers40,41.
Molecular signaling pathways involved in miRNA-mediated regulation of EOC
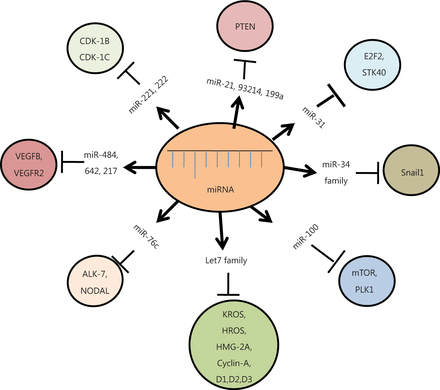
MiRNAs regulate diverse molecular signaling pathways of ovarian cancer pathogenesis by interacting with multiple target genes, and they modulate the functions of these genes across diverse cancer types. The transforming growth factor-β (TGF-β) signaling pathway is one of the best characterized pathways known to play a crucial role in ovarian cancer progression, particularly in epithelial-mesenchymal transition (EMT), through modulation by miR-181a. The key target genes of the TGF-β signaling pathway are the two receptor-regulated SMADs, namely, Smad2 and Smad3, including Smad7. TGF-β also plays a key role in EMT by repressing E-cadherin expression in epithelial cancer cells (the TGF-β/SMAD signaling pathway). Other critical pathways involved in miRNA regulation of EOC include the PI3K/AKT pathway, G-PCR signaling pathway, Wnt/β-catenin pathway, and ERK5 pathway42. Let-7 targets several oncogenes, such as c-Myc, ras, high-mobility group A (HMGA), Janus protein tyrosine kinase (JAK), signal transducer and activator of transcription 3 (STAT3), and NIRF. Another study has suggested that let-7a plays critical roles in tumorigenesis, proliferation, and invasion, possibly by regulating the cell cycle through the NIRF/p53/p21/CDK signaling pathway43. EOC is also characterized by alterations in epidermal growth factor receptor (EGFR), PI3K/AKT/mTOR signaling, and mutations or epigenetic losses of BRCA1/2, PTEN, and TP5344. Aside from the IL-6R-JAK-STAT3 pathway, nuclear factor kappa-B pathway, VEGF pathway, and other pathways have also been reported to regulate various aspects of ovarian carcinoma45 (Figure 2).
Diagrammatic representation of different miRNAs and their target genes. These miRNAs modulate various cellular pathways, either by upregulation or down-regulation of their respective target genes, ultimately causing cancer.
MiRNAs control EMT
MiRNAs are involved in ovarian cancer initiation and progression46. Aberrant expression of miRNAs plays a critical role in cancer etiology and affects cellular functions by modulating the functional genome. Thus, identification of their target genes has gained significant interest for better management of ovarian cancer.
In the pathogenesis of ovarian cancer, transformation of epithelial cells to mesenchymal cells marks the inception of cancer development and invasion. This transformation (EMT) is characterized by a series of reversible events involving molecular reprogramming and phenotypic changes in cells, leading to dedifferentiation of polarized, immotile epithelial cells to motile mesenchymal cells; this dedifferentiation occurs when the level of E-cadherin protein, a class of type-1 transmembrane protein responsible for maintaining identity of epithelial cells, is reduced47. The miR-200 family of miRNAs plays an important role in this transition by targeting ZEB-1 and ZEB-2, the transcriptional repressor of E-cadherin genes48. MiR-200 family members include miRNA-200a, miRNA-200b, miRNA-200c, miR-141, and miR-429. Different members of the miR-200 family influence EMT transition and cancer progression in different manners by affecting multiple target genes and regulatory pathways involved in ovarian carcinoma. For example, a strong and positive correlation between the expression of E-cadherin and miR-200c has been reported in ovarian cancer tissues. Furthermore, the miR-200 family members have also been found to suppress ZEB1 and ZEB2, resulting in subsequent suppression of EMT49. Concomitantly, overexpression of miR-200 a/b/c and/or miR-141 down-regulates ZEB1/2 levels, leading to higher levels of E-cadherin and epithelial phenotype. By contrast, another study reported that ZEB1/2 can also inhibit the expression of E-cadherin by blocking the transcription of miR-200 family members by binding to clusters of the miR-200 promoter. For EMT induction, the cells are activated by external stimuli, such as TGF-β or PDGF-D, which trigger the expression of transcriptional repressor ZEB1/2 and decrease the expression of miR-20050. Nonetheless, miR-200 family members also control the expression of β-tubulin III, as well as its response with paclitaxel-based therapy and progression-free survival in ovarian cancer patients. The down-regulation of the miR-200 family causes increased expression of β-tubulin III, which leads to the development of chemoresistance in ovarian cancer patients51. Reduced expression of miR-200c is associated with recurrence of ovarian cancer. In addition, miR200 overexpression also significantly inhibits ovarian cancer cell invasiveness and metastasis by down-regulating MMP3, possibly through ZEB1/pSMAD352.
MiR-34-a/b/c induced by p53 is down-regulated in ovarian cancer. The p53 mutation or loss-of-function promotes EMT of cancer cells by increasing the expression of Snail1 protein as the miR-34 family of miRNAs suppresses Snail1 activity when it binds to highly conserved 3’-untranslated region in Snail1 and its regulatory molecules, including β-catenin, LEF1, and Axin2. Therefore, mutated p53 down-regulates miR-34a/b/c to maintain the level of Snail1 protein, which is a zinc-finger transcriptional repressor that regulates EMT programs of cancer cells53,54. The possible molecular mechanisms underlying their down-regulation include aberrant promoter methylation of miRNA genes or any alterations in their copy number53.
Oxidative stress produced during the progression of ovarian cancer also affects various aspects of cancer proliferation and therapeutic approach by regulating the expression of miRNAs. MiR-141 and miR-200a modulate the oxidative stress response by targeting p38α. On the one hand, overexpression of these two miRNAs mimics p38α deficiency and increases tumor growth; on the other hand, it improves the response to chemotherapeutic agents in mouse models. High-grade human ovarian adenocarcinomas with increased accumulation of miR-200a show decreased levels of p38α and associated oxidative stress55. Given that the miR200a-dependent stress signature has fair correlation with improved survival of patients in response to treatment, it certainly can serve as a potential biomarker to predict clinical outcome in ovarian cancers. In addition, apart from promoting tumor growth, oxidative stress also sensitizes tumors for treatment and significantly limits the success of antioxidants in clinical trials55.
MiRNAs critically regulate tumor growth, angiogenesis, and cancer cell survival in ovarian cancer
Down-regulation of certain miRNAs promotes tumor growth and survival of cancer cells by inhibiting apoptosis, inducing metastasis/angiogenesis, and developing chemoresistance against many drugs in ovarian carcinoma. Reduced levels of these miRNAs may be insufficient to check the upregulation of several oncogenes, thereby enhancing tumor development. Among the various down-regulated miRNAs, the let-7 family of miRNAs, which targets embryonic genes, such as HMAG2, Mlin-41, and IMP-1, is universally and significantly down-regulated in all expression profiles of ovarian cancer. The early phase of tumorigenesis resembles embryonic development, which involves re-expression of embryonic mesenchymal genes and de-differentiation of cancer cells, and it is caused by down-regulation of the let-7 family of miRNAs36. Let-7 suppresses several oncogenes, such as KRAS, HRAS, and c-MYC, in ovarian cancer35. The reduced level of let-7 in ovarian cancer cells causes upregulation of these oncogenes by enhancing tumor growth and cell survival56, which ultimately leads to poor survival of cancer patients57.
MiR-214 is responsible for the constitutive activation of the PTEN/ AKT pathway, leading to chemoresistance in different types of tumors, including ovarian cancer58. The increased expression of miR-214 is involved in cisplatin resistance in ovarian cancer by down-regulation of PTEN protein and activation of the PI3K/AKT/mTOR pathway, thereby enhancing cell survival58. In addition, miR-21 located on chromosome 17q23 is overexpressed in most cancers and plays a critical role in neoplastic transformation, invasion, and metastatic processes. MiR-21 inhibits apoptosis in cancer cells by targeting PTEN and PDCD4 and activating the AKT pathway59. Similarly, down-regulation of miR-100, a tumor suppressor miRNA, mediates increased sensitivity to everolimus in most ovarian cancer cell lines and tissues primarily through the repression of mTOR-AKT signaling60,61. Ye et al.62 showed that miR-376c promotes cell proliferation, survival, and spheroid formation by suppressing activin receptor-like kinase 7 (ALK7) and its ligand Nodal, which both induce apoptosis in human EOC cells. Chemosensitive and chemoresistant ovarian tumors have shown differential expression of ALK7 and miR-376c. Overexpression of miR-376c blocks cisplatin-induced cell death and enhances cell survival. Another study showed that miRNA-93 stimulates tumor growth and angiogenesis by targeting the integrin-β8 gene63 (Figure 2).
Through a gain-of-function screen using the miRNA mimic library (319 miRNA species) to identify various miRNA species affecting cell proliferation in human EOC cells, A2780 has shown the overexpression of miR-124a, miR-192, miR-193a, and miR-193b and subsequent inhibition of BrdU incorporation in A2780 cells, indicating that these miRNAs affect the cell cycle. Overexpression of miR-193a and miR-193b induces the activation of caspase 3/7, which leads to apoptotic cell death in A2780 cells. MiR-193a also potentially targets the ARHGAP19, CCND1, ERBB4, KRAS, and MCL1 genes and decreases the level of MCL1 protein by binding to 3’-UTR of its mRNA64. MiRNAs also play intricate roles in the development of drug resistance in EOC. Most notably, hsa-miR-152, hsa-miR-200a-3p, hsa-miR-381, and hsa-miR-429 are differentially expressed between drug-sensitive and drug-resistant tissues65. Integrated genomic analyses have predicted the crucial involvement of eight key miRNAs, including miR-506, miR-141, and miR-200a, in regulating 89% of the targets in the network controlling survival of cancer cells in serous ovarian cancer66. Concomitantly, these findings directly implicated the intricate involvement of miRNAs in gene regulation related to tumor growth and cell survival, suggesting that miRNAs could potentially be used for therapeutic purposes in ovarian cancer.
MiRNAs regulate cell cycle in ovarian carcinoma
MiRNAs also include various cell cycle genes as their targets, and modulating their expression plays significant roles in cell cycle progression in various histotypes of ovarian cancers. The let-7 family of miRNAs regulates several cell cycle genes, such as Cyclin-A, D1, D2, D3, and HMG-2A56. MiRNA-31 observed in serous ovarian cancer cell lines and tissues inhibits the expression of multiple cell cycle regulators, such as E2F2 and STK40, which are repressors of p53-mediated transcription67. The enhanced expression of miR-221 and miR-222, which are located on X-chromosome, inhibits apoptosis by down-regulating cell cycle genes, namely, CDK1B (p27) and CDK1C (p57)68. Furthermore, miR-210 is the most prominent miRNA that is consistently stimulated under hypoxic conditions, and it is regulated by the HIF signaling pathway. MiR-210 also regulates the e2f transcription factor 3 (e2f3), a key protein in the cell cycle, as shown by both biocomputational analysis and in vitro studies. MiR-210 down-regulates the expression of E2F in ovarian cancer patients, indicating that miR-210 plays a crucial role in tumor onset as a key regulator of the hypoxic response of tumor cells and provides a link between hypoxia and cell cycle regulation25. More recently, the dysregulation of five miRNAs has been linked to the significant enrichment of the cell cycle, revealing that the MAPK signaling pathway is highly involved in ovarian carcinogenesis; these findings suggested that these miRNAs may function as diagnostic and prognostic biomarkers69 (Figure 2).
MiRNAs as a powerful diagnostic tool for ovarian cancer
Among the various curative and preventive measures considered, early detection has long been the key to successful treatment of multiple life-threatening diseases, including ovarian cancer. Early detection underlies the foundation for better management and intervention of ovarian cancer at its primary stage before it spreads and affects other healthy tissues of the body. The aberrant expression pattern of miRNAs can be a powerful tool to diagnose ovarian cancer in its earliest stages. Strengthening this view, Iorio et al.30 reported the aberrant expression of miRNA in ovarian cancer and found that miR-141 and miR-200a/b/c are the most significantly overexpressed, whereas miR-199, miR140, miR-145, and miR-125b are significantly down-regulated. MiR-200a and miR-200c are overexpressed in all histologic groups, such as serous, endometrioid, clear cell, and mixed carcinoma, whereas miR-200b and miR-141 are endometrioid and serous-specific. A very recent study has also reported significant down-regulated expression of serum miR-145 in patients with malignant ovarian cancer and benign ovarian cancer compared with that in healthy controls, and results suggested that miR-145 can potentially serve as an outstanding biomarker for the detection of ovarian and other human cancers70.
Hypomethylation of miRNA genes is the key epigenetic mechanism for upregulation of miR-21, miR-203, and miR-205 compared with normal ovary30. Another study has reported that upregulation of miRNAs is due to the amplification of miRNA genes in ovarian cancer41. Among the 35 miRNAs analyzed, four miRNAs (miR26, miR182, miR103, and miR26a) were significantly up-regulated, whereas two miRNAs (let-7d and miR-127) were highly down-regulated. These studies pointed out that alterations in copy number and epigenetic silencing of miRNA genes are the two mechanisms of aberrant expression of miRNAs in ovarian cancer41. Furthermore, miR-30c, miR-30d, and miR-30e are frequently up-regulated, whereas miR-493 is down-regulated in ovarian carcinomas compared with that in normal HOSE cell lines. However, miR-30a has been identified as a clear cell-specific miRNA in ovarian cancer71. In a recent study, comparative and detailed miRNA expression profiling was performed in various tissues and malignant ovarian tumors using quantitative real-time PCR (qPCR) to explore the relationship between frequently deregulated miRNAs and clinicopathologic variables, response to therapy, survival, and their association with Her-2/neu status in ovarian carcinomas. This study clearly established that expression levels of the four miRNAs (miR-30c, miR-30d, miR-30e-3p, and miR-370) is significantly higher in ovarian carcinoma than that in benign ovarian tissue, whereas, three miRNAs (miR-181d, miR-30a-3p, miR-532-5p) are significantly different between borderline and ovarian carcinoma. Most importantly, miR-370, which is highly up-regulated in early stage of ovarian cancer, can be used as a biomarker for early detection of ovarian cancer. In addition, the down-regulation of miR-30c, miR-30d, miR-30e-3p, and miR-532-5p is associated with the overexpression of Her2/neu oncogene, whereas the overexpression of miR-30a-3p and miR-181d is associated with well-differentiated carcinomas (grade 1) compared with poorly differentiated carcinomas (grade 3), showing the diagnostic potential of multiple miRNA species for differential staging and grading of ovarian carcinoma72.
Aside from the expression profiling of miRNA in different tissues, blood-based serum and plasma miRNA expression is also well documented and may be used as surrogate non-invasive biomarkers. For example, miRNA expression profiling in serum and tumor-derived exosomes from the same patients revealed that eight miRNAs (miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, and miR-214) are similar, both in tissue and tumor-derived exosomes73. Furthermore, a recent study also showed the potential of miRNAs as stable blood-based noninvasive biomarkers using serum miRNAs from EOC patients. This study reported the differential expression of eight miRNAs between cancer patients and normal controls. Among these miRNAs, five miRs, namely, miR-21, miR-92, miR-93, miR-126, and miR-29a, are significantly overexpressed, whereas three miRs, namely, miR-155, miR-127, and miR-99b, are significantly down-regulated in serum of cancer patients than their matching controls74. MiRNAs 21, 92, and 93 are overexpressed in patients with normal CA125 levels, further suggesting that miRNAs can be used for early diagnosis of ovarian cancer75. The above study strongly suggested that expression profiling of circulatory tumor-derived miRNAs can potentially be used as surrogate biomarkers for EOC detection, as well as screening of asymptomatic populations. In addition, a recent study observed the upregulation of serum miR-221 in patients with EOC compared with healthy controls, and the expression level of serum miR-221 was found to be significantly associated with stage and grade of tumor. Serum miR-221 may function as a novel diagnostic and prognostic marker and it may also serve as a potential therapeutic target in EOC treatment. MiR-200a and miR-449b are highly overexpressed in stage I than in stage III ovarian carcinoma, so they may facilitate the detection of early stage EOC76 (Table 1). ZEB1, a miR-200c target gene, does not seem to be regulated in stage I EOC, suggesting that MiR-200c may play a different role in stage I EOC76. A recent study suggested that plasma miR-205 and let-7f are strong biomarkers for the detection of ovarian cancer complementing CA-125. In addition, let-7f may also serve as a reliable predictive biomarker for ovarian cancer prognosis77. Moreover, the above studies collectively suggested that aberrant miRNA expression patterns in tissues, serum, and tumor-derived exosomes form patients with ovarian carcinoma and control samples have significant great therapeutic importance for EOC and strongly indicate that they can be used as diagnostic biomarkers for ovarian cancer patients (Table 2).
Role of various up-regulated and down-regulated miRNAs in ovarian cancer diagnosis, prognosis, and chemoresistance
Summary of the roles of miRNAs associated with diagnosis and prognosis, type of tissue taken, number of sample taken, and discovery platform used
MiRNAs as potential prognosis marker of ovarian cancer
Several studies have reported that aberrant miRNA expression can be used as a prognostic marker to check the disease outcome during the treatment regimen. For example, in ovarian cancers, loss of let-7 and up-regulation of HMGA2 are significantly associated with unfavorable prognosis29. Thus, the ratio of HMGA2 to let-7 has been used for prognosis of ovarian cancer; higher ratio of HMGA2/let-7 exhibits poor survival (<10%) compared with a lower ratio (40%)29. In addition, let-7 used as a prognostic marker in ovarian cancer is down-regulated in chemotherapy-resistant patients and associated with shorter progression-free survival, so it may also serve as a biomarker to observe disease outcome and survival in ovarian cancer patients34. The prognostic value of miRNAs has also been evaluated by expression profiling in 55 ovarian cancer patients, and their subsequent expression levels are correlated with the disease outcome83. In this study, miR-200a showed significant association with cancer survival, and overexpression of miR-200a predicted favorable disease outcome among the 96 miRNAs analyzed83. However, another study showed that overexpression of miR-200a exhibits poor prognosis84. These contradictory results forced the requirement of large prospective studies to establish miR-200 family members as prognostic markers for ovarian cancer. MiR-9 and miR-223 are correlated with the recurrence of ovarian cancer and may be used as prognostic markers for disease outcome in ovarian cancer82. Recently, miR-30d has been observed to be up-regulated in patients with platinum-based chemosensitive response than in patients with chemo-resistant response. In this study, miR-30d was also observed to be down-regulated in recurrent ovarian cancer patients72. Furthermore, miRNA expression patterns were analyzed using tissues and cell lines of ovarian cancer; miR-221 was observed to be the most consistently overexpressed and the let-7 family members were the most frequently under-expressed miRNAs in ovarian cancers85. These finding suggested that these miRNAs may serve as potential biomarkers for disease outcome.
Another study analyzed the promoter methylation of let-7a-3 using 214 clinical samples of EOC86 and found that hypermethylation limits the risk of deaths in ovarian cancer patients. The methylation pattern of let-7a-3 is inversely correlated with the expression of IGF-II, and let-7a-3 overexpression in tumors is associated with poor prognosis86. Since then, a number of studies have shown the association between aberrant methylation and alterations in chromatin structure with the deregulated expression of miRNAs and consequent development of cancer. One study showed the differential expression patterns of miRNAs in serous ovarian cancer and control, and revealed that miR-21 is frequently up-regulated and miR-125b is frequently down-regulated miRNAs, in most cases84. The study also showed that miRNA expression between normal and malignant ovaries varies and can be used to differentiate the histotypes.
Overexpression of miR-23a, miR-27a, miR-21, and miR24-2 is significantly associated with poor prognosis in patients with progressive disease during first-line chemotherapy. Overexpression of miR-378 is associated with chemosensitization, whereas its under-expression results in chemoresistance among patients treated with platinum-based chemotherapy75,78. Programmed cell death 4 (PDCD4), an important tumor suppressor gene, influences transcription and translation of multiple genes through modulation of different signal transduction pathways. MiRNA-182, which directly targets PDCD4, is up-regulated in ovarian cancer tissues and cell lines. Furthermore, miR-182 reduces the chemosensitivity of ovarian cancer cells, which may be attributed to its anti-apoptotic activity to cisplatin and Taxol (Table 2)80.
The down-regulated expression of miRNA-22 (miR-22) in EOC is linked with overall survival and progression-free survival, so it may serve as an efficient prognostic factor for EOC patients81. Detection of miR-92 levels in the serum is an indicator of clinical stage of tumor, and it can be used as a new tumor biomarker in the diagnosis and assessment of prognosis of EOC87. MiRNA expression profiles also influence chemoresistance in serous epithelial ovarian carcinomas. Three miRNAs (miR-484, -642, and -217) are reportedly critical in predicting chemoresistance of these tumors. Among them, miR-484 is involved in modulating tumor vasculature by regulating the VEGFB and VEGFR2 pathways. Collectively, these three miRNAs can classify the response of ovarian cancer patients to chemotherapy in a large multicenter cohort, whereas miR-484 is involved in the control of tumor angiogenesis, indicating an option in treating patients suffering from serous epithelial ovarian carcinomas79.
Future prospective
Incessant studies from the last two decades have shown that miRNAs are critical regulators of ovarian cancer and play diverse roles from EMT to cancer survival, in cell cycle regulation, for diagnosis and prognosis of cancer, and in therapeutic response to drugs and development of chemoresistance. Citing their diverse roles in each and every stage of cancer pathophysiology as evidenced by miRNA expression profiling, miRNAs can be used as ideal and highly sensitive biomarkers for different subtypes of ovarian cancers. Despite inherent technological limitations and constraints in designing studies to establish miRNAs as clinical molecular biomarkers, they still offer great hope for diagnosis, prognosis, and prediction of EOC because they resemble most of the characteristics of an ideal biomarker. Interestingly, the inherent stabilities and resilience of miRNAs in tumor tissues and blood samples are important for their applications as a reliable and rapid biomarker. The study of gene expression profiling and its clinical associations has been identified in small samples; hence, the translation of miRNAs into clinical biomarkers demands independent studies at larger scales. In addition, studies focusing on tumor-specific microvesicles are required to provide insights into the biological roles of circulating miRNAs. Apart from well-noted diagnostic and prognostic values, miRNAs also provide a potential treatment option for ovarian cancer by inhibiting the expression of pathologically relevant genes, which is mainly mediated through post-transcriptional gene silencing. Considering these facts and given the higher sensitivities and specificities of miRNAs compared with other currently used clinical biomarkers, they hold potential for use as substantial clinical biomarkers for early stage detection, monitoring progression, and therapeutics of ovarian cancer.
Conclusion
Extensive studies from the last two decades have clearly suggested that miRNAs are critical regulators of gene expression and cell cycle machinery, thereby maintaining the differentiation status in ovarian carcinoma cells. Any deregulation in miRNA expression may trigger cancer development, cancer aggressiveness (e.g., low grade and high grade tumors), chemoresistance, migration, and metastasis. In addition, miRNA expression profiling in healthy and benign tumors may entirely differ from that in cancerous cells. Thus, expression patterns of miRNA population may serve as a prognostic and reliable biomarker for early detection, which may help in effective and improved management of ovarian carcinoma. Furthermore, identification of miRNAs associated with the different histotypes of ovarian cancer, resistance to drugs, cell invasion, and metastasis may help in designing better strategies for more individualized and efficient treatment regimens for ovarian cancer. Most of the currently available options to treat cancers are site-directed because of the limited current knowledge about disease progression and the fact that it is the most effective treatment strategy to execute. Given that identification of the primary site of tumor poses many clinical challenges and has the most significant role in treating cancers, miRNA expression profiles may improve the diagnosis of tumors from unknown primary origin, which would help in ensuring management plan and lead to better survival of ovarian cancer patients. In addition, tumor-specific and blood-based circulating miRNAs may improve cancer diagnosis and prognosis. Fortunately, several promising miRNAs have already been explored as non-invasive biomarkers for different tumor entities. At this juncture, we have only started to evaluate the role of miRNAs as clinical biomarkers for early stage diagnosis of ovarian cancer. However, before drawing any final conclusion and using miRNAs as more reliable and non-invasive biomarkers for the detection and treatment of ovarian cancer, future studies comprising large sample sets with well-supported validation through long-term clinical data must be conducted. Finally, before any clinical application of miRNAs, high-throughput optimization is critical to enhance detection under different stages and conditions of ovarian cancer pathogenesis, as well as for their reliable efficacy in cancer therapy.
Acknowledgements
We gratefully acknowledge the assistance of Dr. Manjunatha Prabhu, Endocrinology Division, CDRI, Lucknow, in the design of the manuscript and Akancha Pandey, Department of Obstetrics and Gynecology, KGMU, for the preparation of the review. We also acknowledge the ICMR New Delhi for financial support (Grant No. 3/2/2/136/2012/NCD-III).
Footnotes
Conflict of interest statement No potential conflicts of interest are disclosed.
- Received April 2, 2015.
- Accepted June 15, 2015.
- Copyright: © 2015, Cancer Biology & Medicine
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY) 4.0, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1.↵
- 2.↵
- 3.↵
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.
- 13.↵
- 14.↵
- 15.
- 16.
- 17.
- 18.↵
- 19.↵
- 20.
- 21.
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.↵
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.↵
- 78.↵
- 79.↵
- 80.↵
- 81.↵
- 82.↵
- 83.↵
- 84.↵
- 85.↵
- 86.↵
- 87.↵